|
| |
|
|
|
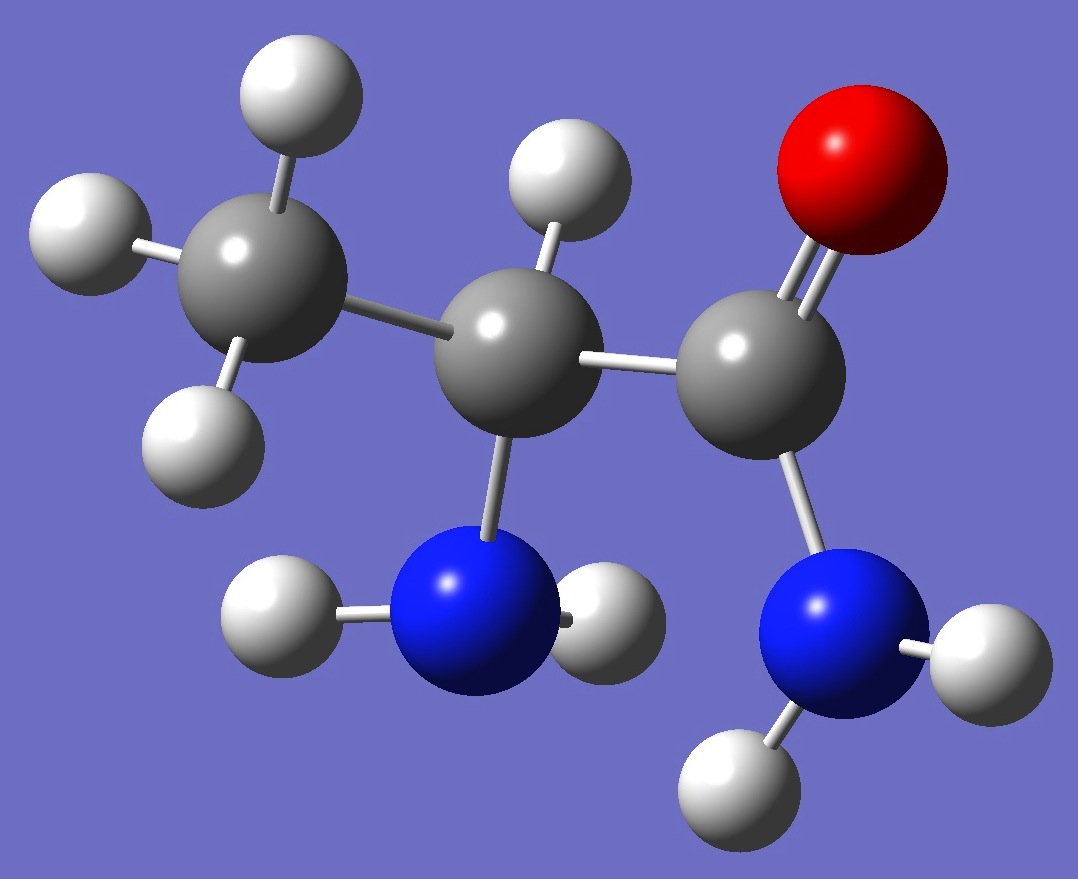
Table 3. Alaninamide.
Structure parameters (Å and
degrees).
|
| |
|
|
|
|
|
|
|
|
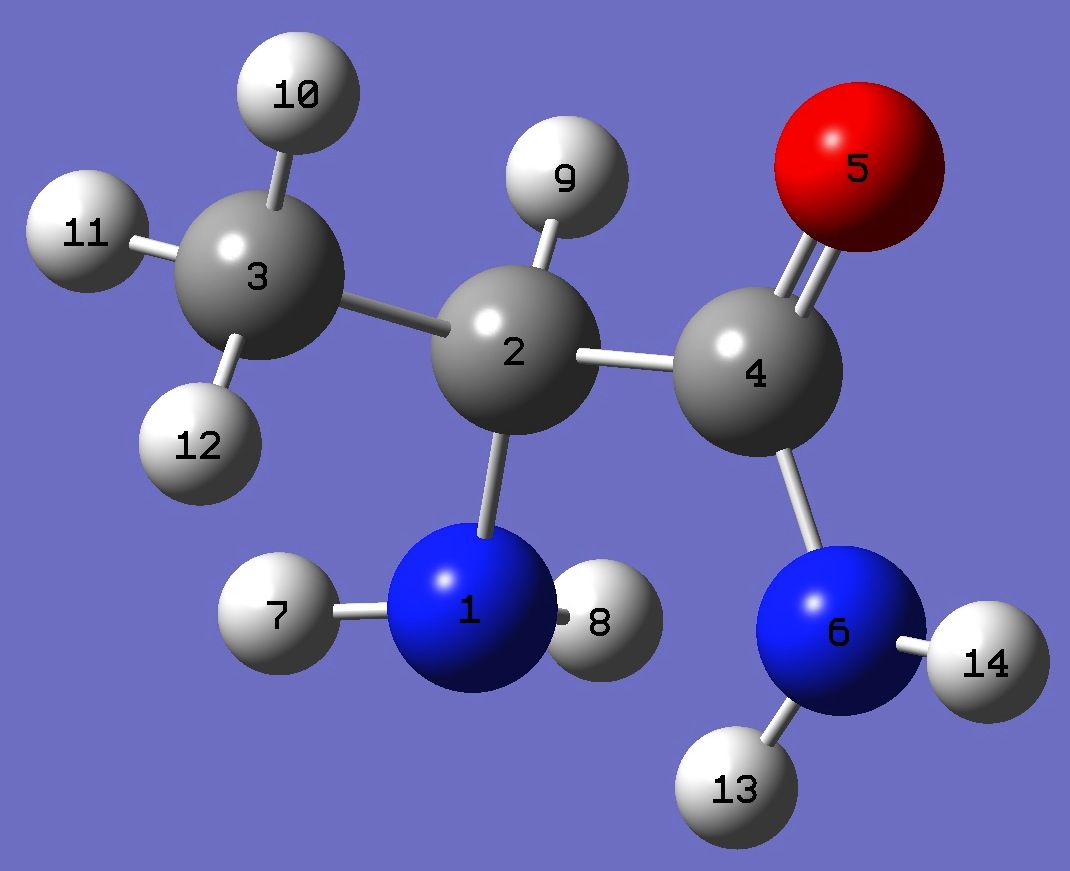
N
C,1,B1
C,2,B2,1,A1
C,2,B3,1,A2,3,D1,0
O,4,B4,2,A3,1,D2,0
N,4,B5,2,A4,1,D3,0
H,1,B6,2,A5,4,D4,0
H,1,B7,2,A6,4,D5,0
H,2,B8,1,A7,3,D6,0
H,3,B9,2,A8,1,D7,0
H,3,B10,2,A9,1,D8,0
H,3,B11,2,A10,1,D9,0
H,6,B12,4,A11,2,D10,0
H,6,B13,4,A12,2,D11,0
|
|
|
|
|
|
|
|
|
|
|
| Best-Fit * |
ropt |
|
|
|
|
|
B1=1.48284
B2=1.51962
B3=1.52768
B4=1.25784
B5=1.33077
B6=1.01668
B7=1.01657
B8=1.10213
B9=1.09363
B10=1.09581
B11=1.09590
B12=1.00976
B13=1.00781
A1=109.656
A2=109.349
A3=118.970
A4=117.226
A5=108.805
A6=109.265
A7=113.868
A8=109.737
A9=110.861
A10=109.852
A11=117.579
A12=117.318
D1=121.852
D2=167.405
D3=-21.102
D4=171.214
D5=-73.763
D6=-121.594
D7=175.760
D8=-64.135
D9=55.850
D10=-3.478
D11=-159.621
|
B1=1.4640073
B2=1.52378867
B3=1.53078929
B4=1.21834716
B5=1.35063777
B6=1.01434923
B7=1.01466118
B8=1.09813415
B9=1.09057229
B10=1.09408973
B11=1.09395509
B12=1.00950015
B13=1.00621127
A1=110.12776684
A2=111.43520268
A3=121.01736175
A4=114.71141281
A5=110.08718563
A6=110.48578466
A7=113.15106644
A8=109.97526796
A9=110.67739522
A10=110.50843693
A11=118.47598753
A12=119.15029805
D1=120.4281597
D2=165.60127299
D3=-15.29168762
D4=160.29119012
D5=-82.45345113
D6=-121.46899311
D7=175.01671191
D8=-65.1733097
D9=54.92649236
D10=-2.78705813
D11=-174.66751537
|
|
|
|
|
|
* Generated here from best-fit atomic coordinates given in Ref. [1].
|
|
|
|