|
| |
|
|
|
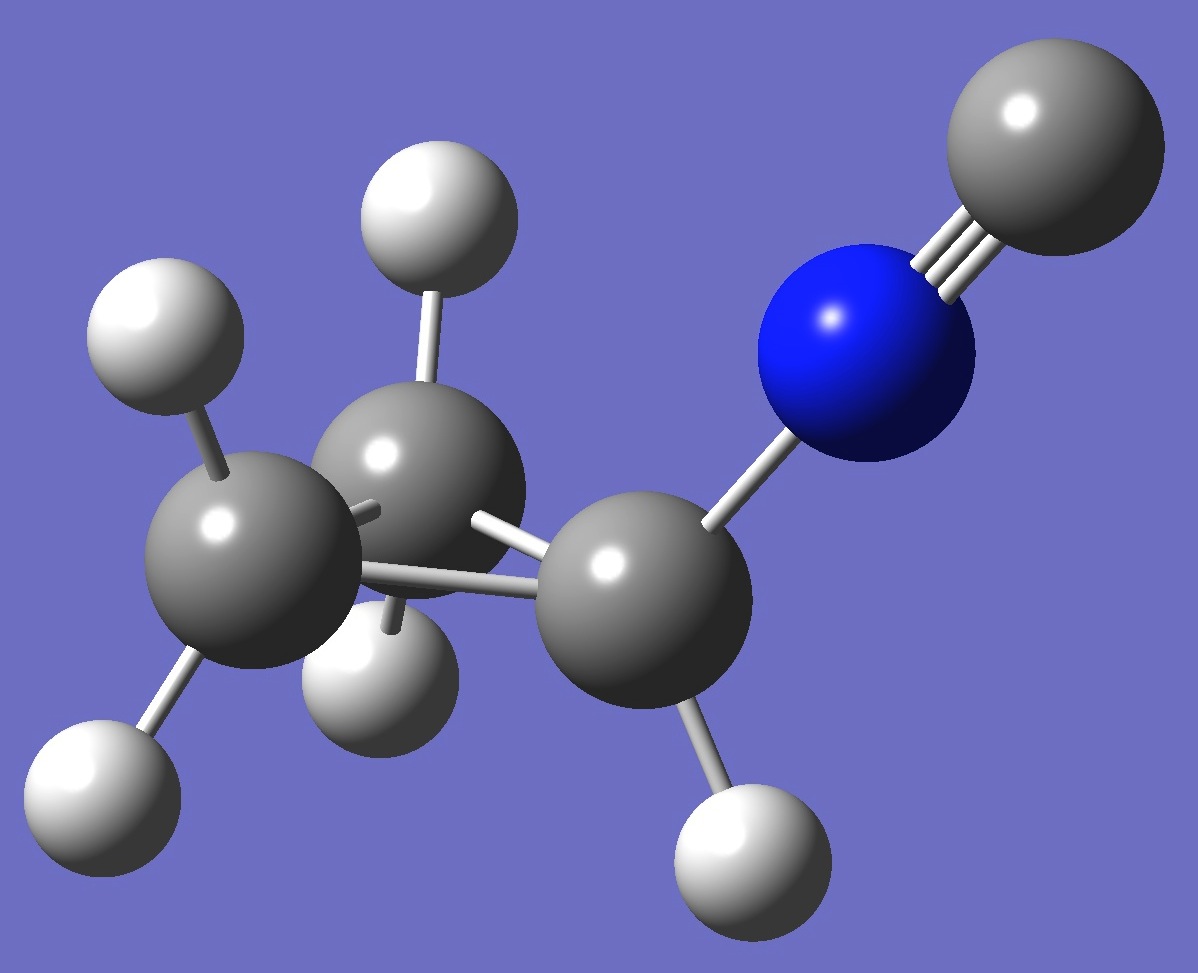
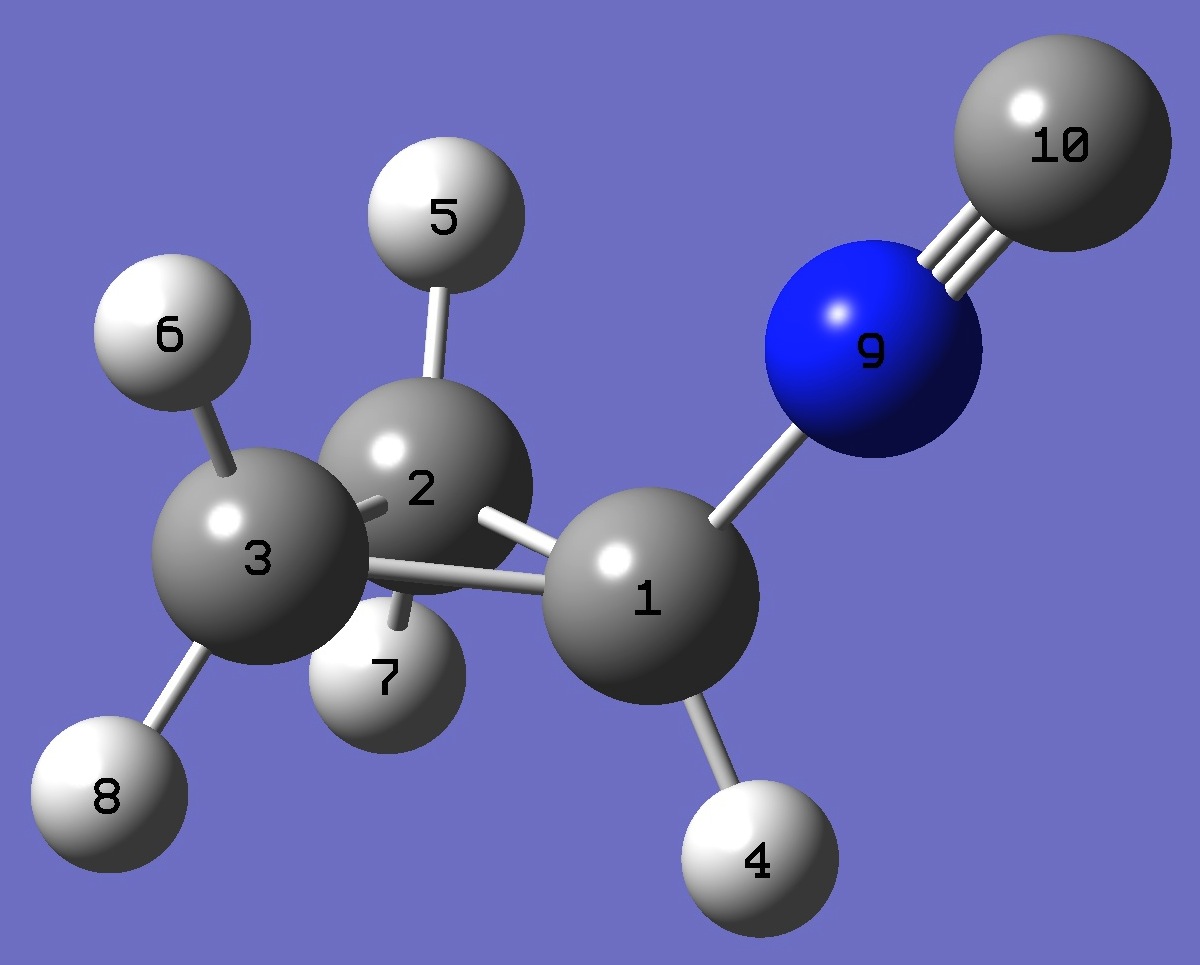
| Table 2. Molecular
optimized structure parameters: ropt(1) = PBE1PBE/6-31G(3d,3p) and ropt (2) = PBE1PBE/6-311+G(3d,3p) (Å
and degrees). |
| |
|
|
|
|
C
C,1,B1
C,1,B2,2,A1
H,1,B3,2,A2,3,D1,0
H,2,B4,1,A3,3,D2,0
H,3,B5,1,A4,2,D3,0
H,2,B6,1,A5,3,D4,0
H,3,B7,1,A6,2,D5,0
N,1,B8,2,A7,3,D6,0
C,9,B9,1,A8,3,D7,0
|
|
|
|
|
|
|
ropt(1) |
ropt(2) |
|
|
B1=1.50239231
B2=1.50239231
B3=1.0845076
B4=1.08353945
B5=1.08353945
B6=1.08384192
B7=1.08384192
B8=1.39368335
B9=1.17143198
A1=59.77857918
A2=118.30324052
A3=116.54823659
A4=116.54823659
A5=117.06634501
A6=117.06634501
A7=118.86929516
A8=179.62391104
D1=108.03074862
D2=108.30484966
D3=-108.30484966
D4=-109.5097824
D5=109.5097824
D6=-108.4750738
D7=145.31636934
|
B1=1.50163845
B2=1.50163845
B3=1.0819369
B4=1.0810289
B5=1.0810289
B6=1.0815039
B7=1.0815039
B8=1.39328142
B9=1.16702295
A1=59.83136681
A2=118.32297758
A3=116.50054017
A4=116.50054017
A5=116.98277346
A6=116.98277346
A7=118.80827445
A8=179.64641339
D1=108.06605261
D2=108.22103568
D3=-108.22103568
D4=-109.43614583
D5=109.43614583
D6=-108.44724993
D7=145.30785944
|
|
|
|
|
|
|