|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3SSCH3
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Sulfur
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
|
in Dimethyl Disulfide |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The complete nqcc tensor for 33S in CH333S32SCH3 was determined by Hartwig et al. [1]. A molecular ro
structure was derived by Sutter et al. [2]. Calculation of the
nqcc tensor was made here on this structure, and on this structure but
with the geometry of the methyl groups determined ab initio as described below.
|
|
|
Calculated and experimental nqcc
tensors are compared in Tables 1 and 2. Structure parameters are
given in Table 3, atomic coordinates in Table 4. |
|
|
|
|
|
|
|
|
|
|
|
|
In Tables 1 and 2, subscripts a,b,c refer to the principal axes of the inertia
tensor, subscripts x,y,z to the principal axes of the nqcc tensor. Ø (degrees)
is the angle between its subscripted parameters. ETA = (Xxx
- Xyy)/Xzz. |
|
|
RMS is the root mean square
difference between calculated and experimental nqcc's (percentage of
average experimental nqcc). RSD are the residual standard deviations
of calibration of the B3LYP/6-311G(3df,3p) and B3LYP/TZV+(3df,3p) models for calculation of
the nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 1. 33S nqcc's in CH333S32SCH3 (MHz). Calculation was made with the B3LYP/6-311G(3df,3p) model on the ro and ro/ropt structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. ro
|
|
Calc. ro/ropt |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
30.41 |
- |
29.94 |
- |
29.6475(42) |
|
|
Xbb |
|
13.26 |
|
12.86 |
|
12.807(31) |
|
|
Xcc |
|
17.15 |
|
17.08 |
|
16.840(26) |
|
|
Xab* |
- |
21.52 |
- |
21.90 |
|
20.2(13) |
|
|
Xac* |
|
15.63 |
|
15.61 |
|
18.3(12) |
|
|
Xbc* |
- |
24.59 |
- |
24.73 |
|
24.43(17) |
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.54 (2.8 %) |
0.22 (1.1 %) |
|
|
|
RSD |
|
0.39 (1.7 %) |
0.39 (1.7 %) |
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
- 8.70 |
|
- 8.97 |
|
- 9.52(32) |
|
|
Xyy |
- |
39.84 |
- |
39.69 |
- |
39.2(11) |
|
|
Xzz |
|
48.54 |
|
48.66 |
|
48.7(12) |
|
|
ETA |
|
0.641 |
|
0.631 |
|
|
|
|
Øz,n** |
|
0.5 |
|
0.3 |
|
|
|
|
Øx,bi** |
|
12.1 |
|
11.4 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Here and in Table 2 below,
|
|
|
* the algebraic signs of the off-diagonal nqcc's correspond to the atomic
coordinate given in Table 4. The experimental off-diagonal
components are absolute values.
|
|
|
** the z-principal axis makes an angle of Øz,n with the normal (n) to the C(2)33S(1)S(2) plane, the x-axis makes an angle of Øx,bi with the bisector (bi) of the C(2)33S(1)S(2) angle. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 2. 33S nqcc's in CH333S32SCH3 (MHz). Calculation was made with the B3LYP/TZV+(3df,3p) model on the ro and ro/ropt structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. ro
|
|
Calc. ro/ropt |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
30.46 |
- |
29.98 |
- |
29.6475(42) |
|
|
Xbb |
|
13.36 |
|
12.96 |
|
12.807(31) |
|
|
Xcc |
|
17.10 |
|
17.03 |
|
16.840(26) |
|
|
Xab* |
- |
21.50 |
- |
21.88 |
|
20.2(13) |
|
|
Xac* |
|
15.78 |
|
15.76 |
|
18.3(12) |
|
|
Xbc* |
- |
24.60 |
- |
24.74 |
|
24.43(17) |
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.58 (3.0 %) |
0.24 (1.2 %) |
|
|
|
RSD |
|
0.35 (1.5 %) |
0.35 (1.5 %) |
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
- 8.74 |
|
- 9.01 |
|
- 9.52(32) |
|
|
Xyy |
- |
39.89 |
- |
39.74 |
- |
39.2(11) |
|
|
Xzz |
|
48.62 |
|
48.74 |
|
48.7(12) |
|
|
ETA |
|
0.641 |
|
0.630 |
|
|
|
|
Øz,n** |
|
0.4 |
|
0.3 |
|
|
|
|
Øx,bi** |
|
12.4 |
|
11.7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
For the ro structure, all CH bond lengths and SCH angles are assumed equal. For the ro/ropt structure,
methyl geometries are given by MP2/6-31G(d,p) optimization, with CH
bond lengths corrected using r(CH) = 1.001*ropt(CH) [3]. |
|
|
| |
|
|
|
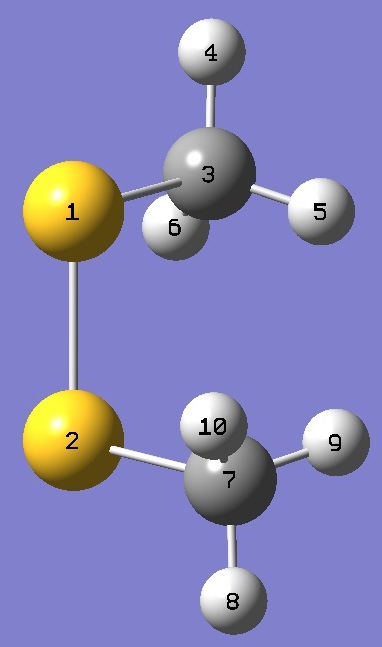
| Table 3. Molecular structure parameters, ro and ro/ropt (Å and degrees). Complete structures are given here in Z-matrix format. |
| |
|
|
|
 |
|
ro |
ro/ropt |
|
|
|
| SS |
2.038 |
2.038 |
| SC |
1.810 |
1.810 |
| SSC |
102.8 |
102.8 |
| CSSC |
84.7 |
84.7 |
| CH(4,8) |
1.097 |
1.090 |
| CH(5,9) |
1.097 |
1.088 |
| CH(6,10) |
1.097 |
1.087 |
| SCH(4,8) |
108.9 |
106.6 |
| SCH(5,9) |
108.9 |
111.5 |
| SCH(6.10) |
108.9 |
110.8 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
| Table 4. CH333S32SCH3 Atomic coordinates, ro/ropt. |
|
|
| |
|
|
|
|
|
|
|
|
|
|
a (Å) |
|
b (Å) |
|
c (Å) |
|
|
|
|
|
|
|
|
|
33S |
|
0.9099 |
- |
0.4308 |
- |
0.4326 |
|
32S |
- |
0.9264 |
- |
0.4272 |
|
0.4514 |
|
C |
|
1.7826 |
|
0.8863 |
|
0.4503 |
|
H |
|
2.7961 |
|
0.9120 |
|
0.0497 |
|
H |
|
1.3157 |
|
1.8547 |
|
0.2833 |
|
H |
|
1.8239 |
|
0.6754 |
|
1.5158 |
|
C |
- |
1.8078 |
|
0.8643 |
- |
0.4602 |
|
H |
- |
2.8214 |
|
0.8921 |
- |
0.0603 |
|
H |
- |
1.3473 |
|
1.8392 |
- |
0.3146 |
|
H |
- |
1.8475 |
|
0.6298 |
- |
1.5209 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1,2-Dithiin |
Disulfane |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] H.Hartwig, U.Kretschmer, and H.Dreizler,
Z.Naturforsch. 50a,131(1995).
|
|
|
[2] D.Sutter, H.Dreizler,
and H.D.Rudolph, Z.Naturforsch. 20a,1676(1965). |
|
|
[3] J.Demaison and G.Wlodarczak, Struct.Chem. 5,57(1994). |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Sulfur |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3SSCH3.html |
|
|
|
|
|
|
Last
Modified 18 April 2006 |
|
|
|
|
|
|
|
|
|
|

