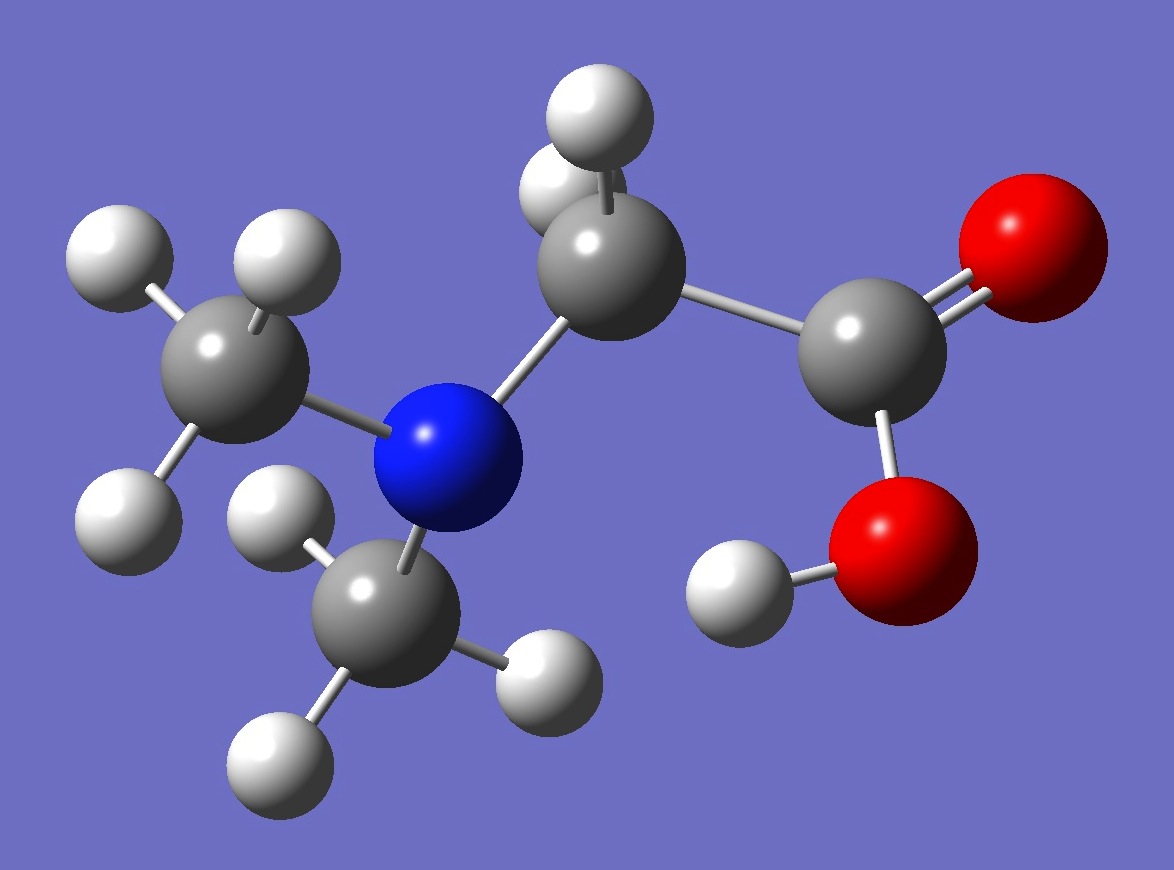
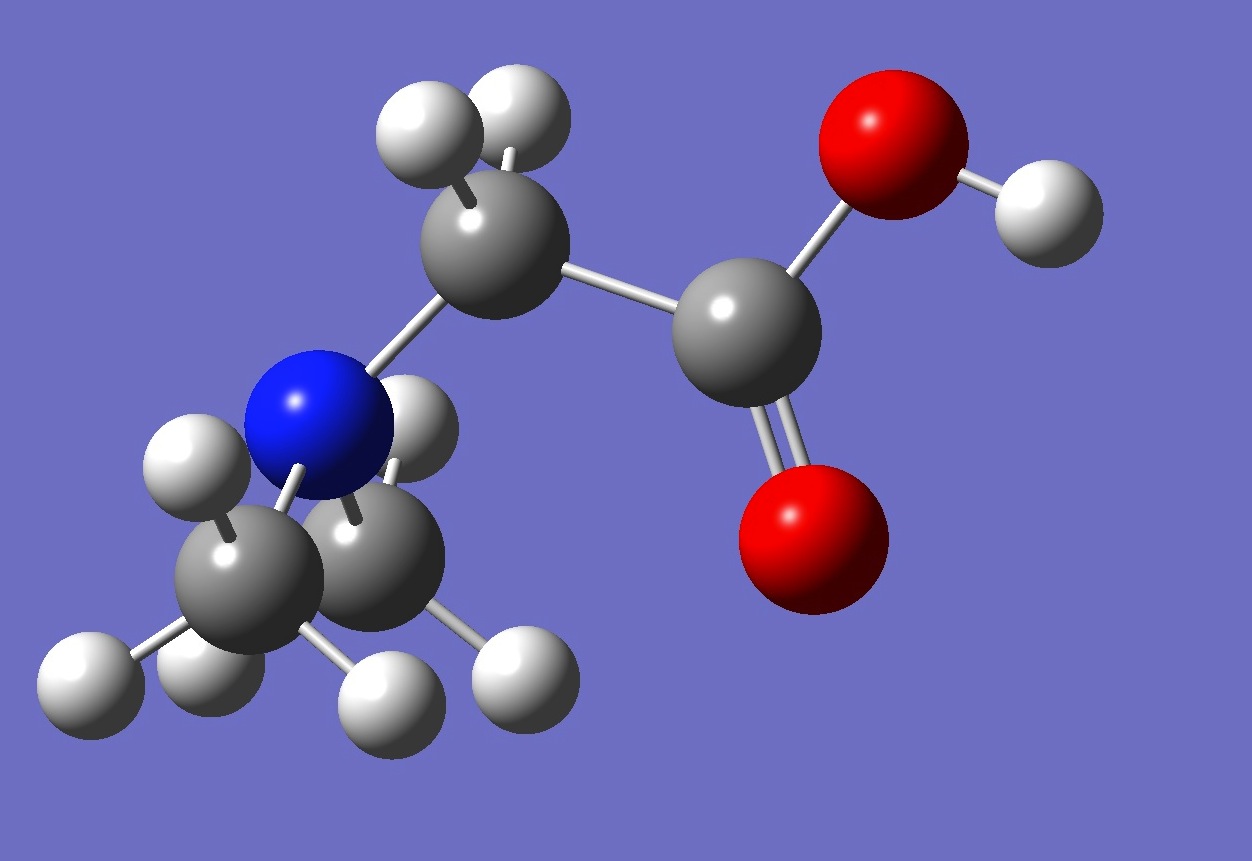
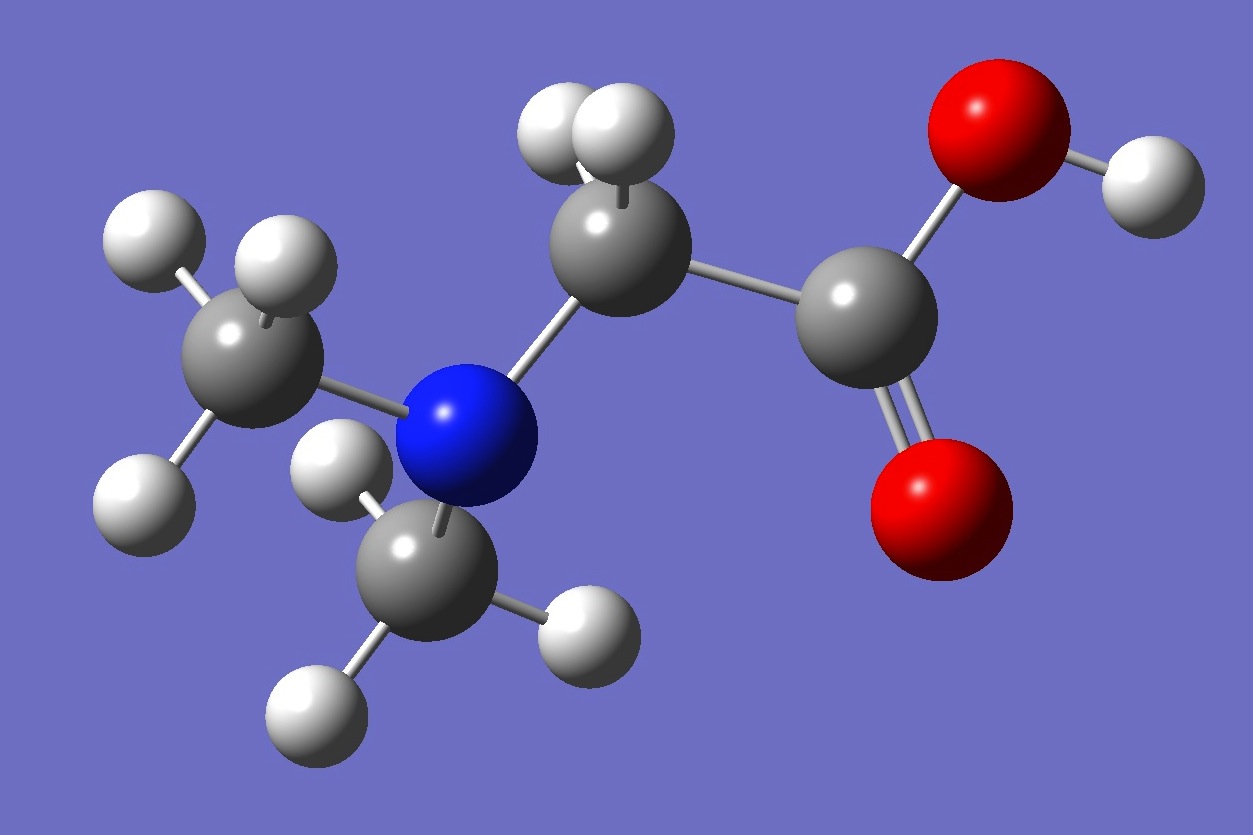
| |
||||||||
| Table
1. 14N nqcc's in Conformer 1 (MHz).
Calculation was made on (1) B3P86/6-31G(3d,3p) and (2)
mPW1PW91/6-31G(3d,3p) optimized structures. |
||||||||
| |
||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
||||||
| Xaa | 1.845 |
1.861 |
1.9637(12) |
|||||
| Xbb | 0.637 |
0.645 |
0.5586(23) |
|||||
| Xcc | - |
2.483 |
- |
2.506 |
- |
2.5223(23) |
||
| Xab | 1.281 |
1.276 |
||||||
| Xac | 1.985 |
1.981 |
||||||
| Xbc | - |
3.042 |
- |
2.506 |
||||
| |
||||||||
| RMS |
0.085 (5.1 %) |
0.078 (4.7 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.477 |
2.487 |
||||||
| Xyy | 2.662 |
2.670 |
||||||
| Xzz | - |
5.140 |
- |
5.157 |
||||
| |
||||||||
| |
||||||||
| Table 2. 14N nqcc's in Conformer 2 (MHz).
Calculation was made on (1) B3P86/cc-pVDZ and (2)
mPW1PW91/cc-pVDZ optimized structures. |
||||||||
| |
||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
||||||
| Xaa | 0.580 |
0.593 |
0.5689(16) |
|||||
| Xbb | - |
3.336 |
- |
3.360 |
- |
3.3212(24) |
||
| Xcc | 2.756 |
2.767 |
2.5723(24) |
|||||
| Xab | 4.109 |
4.113 |
||||||
| |
||||||||
| RMS |
0.013 (0.57 %) |
0.028 (1.3 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.756 |
2.767 |
||||||
| Xyy | 3.174 |
3.180 |
||||||
| Xzz | - |
5.930 |
- |
5.947 |
||||
| |
||||||||
| |
||||||||
| Table 3. 14N nqcc's in Conformer 3 (MHz).
Calculation was made on (1) B3P86/6-31G(3d,3p) and (2)
mPW1PW91/6-31G(3d,3p) optimized structures. |
||||||||
| |
||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
||||||
| Xaa | 2.692 |
2.687 |
2.6668(13) |
|||||
| Xbb | 1.747 |
1.721 |
1.7194(18) |
|||||
| Xcc | - |
4.439 |
- |
4.408 |
- |
4.3862(18) |
||
| Xab | - |
0.652 |
- |
0.667 |
||||
| Xac | 1.472 |
1.490 |
||||||
| Xbc | 2.578 |
2.620 |
2.82(15) | |||||
| |
||||||||
| RMS |
0.037 (1.3 %) |
0.017 (0.58 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.646 |
2.655 |
||||||
| Xyy | 3.034 |
3.037 |
||||||
| Xzz | - |
5.680 |
- |
5.692 |
||||
| |
||||||||
| |
||||
| Table 4. N,N-Dimethylglycine, Rotational Constants (MHz). |
||||
| B3P86/ |
mPW1PW91/ |
|||
| 6-31G(3d,3p) |
6-31G(3d,3p) | Expt. [1] | ||
| Conformer 1 | A |
5121. |
5125. |
5086.50998(55) |
| B |
1613. |
1614. |
1606.56158(115) |
|
| C |
1543. |
1544. |
1536.58238(114) |
|
| cc-pVDZ |
cc-pVDZ | |||
| Conformer 2 | A |
4735. |
4744. |
4714.17739(82) |
| B |
1696. |
1695. |
1691.6780(50) |
|
| C |
1681. |
1681. |
1678.0189(49) |
|
| 6-31G(3d,3p) | 6-31G(3d,3p) | |||
| Conformer 3 |
A |
5290. |
5294. |
5256.19735(23) |
| B |
1633. |
1631. |
1625.134199(99) |
|
| C |
1389. |
1391. |
1382.871741(101) |
|