| Table 1. 14N nqcc's in N-Ethylformamide (MHz). Calculation was made on the rs/ropt structure (the derivation of which is briefly discussed below), and on an ropt structure given by HF/aug-cc-pVTZ optimization. | ||||||||
| Calc rs/ropt | Calc ropt | Expt. [1] | ||||||
| Xaa | 1.389 | 1.384 |
1.3794(13) | |||||
| Xbb | 0.845 | 0.795 |
0.8171(20) | |||||
| Xcc | - | 2.234 | - |
2.179 |
- | 2.1965(20) | ||
| Xab |
- | 0.735 | - |
0.592 |
||||
| Xac |
- | 1.536 | - |
1.569 |
||||
| Xbc |
- | 2.318 | - |
2.265 |
||||
| RMS | 0.027 (1.9 %) | 0.017 (1.1 %) |
||||||
| RSD | 0.030 (1.3 %) | 0.030 (1.3 %) | ||||||
| Xxx | 1.896 | 1.740 |
||||||
| Xyy | 2.117 | 2.183 |
||||||
| Xzz | - | 4.013 | - |
3.923 |
||||
| ETA | 0.055 | 0.113 |
||||||
| Table 2. Molecular structure parameters, rs/ropt and ropt = HF/aug-cc-pVTZ optimization. (Å and degrees). The rs/ropt structure was derived with substitution parameters were held fixed, while hydrogen atom parameters were determined by partial optimization at the MP2/6-311+G(d,p) level of theory. | ||
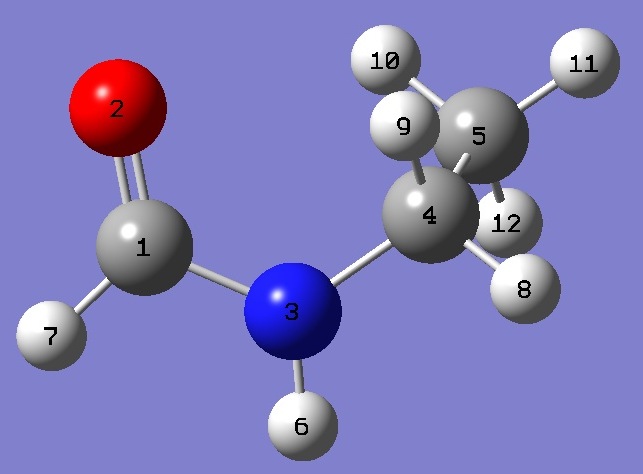 |
C O,1,B1 N,1,B2,2,A1 C,3,B3,1,A2,2,D1,0 C,4,B4,3,A3,1,D2,0 H,3,B5,1,A4,2,D3,0 H,1,B6,2,A5,3,D4,0 H,4,B7,3,A6,1,D5,0 H,4,B8,3,A7,1,D6,0 H,5,B9,4,A8,3,D7,0 H,5,B10,4,A9,3,D8,0 H,5,B11,4,A10,3,D9,0 |
|
| rs/ropt | ropt | |
| B1=1.2173 B2=1.3757 B3=1.439 B4=1.5155 B5=1.00888616 B6=1.10448059 B7=1.09430186 B8=1.09332725 B9=1.0924911 B10=1.09445652 B11=1.09414336 A1=124.02 A2=121.27 A3=113.2 A4=117.88106556 A5=123.4769532 A6=107.84966238 A7=107.38619512 A8=110.08425781 A9=110.50669638 A10=110.83813921 D1=-1.99 D2=-82.17 D3=-174.99295103 D4=179.3530347 D5=154.94222193 D6=39.32471617 D7=59.90859603 D8=179.63296236 D9=-60.29538215 |
B1=1.19185096 B2=1.34234003 B3=1.45100172 B4=1.52249681 B5=0.99027358 B6=1.09118643 B7=1.08263965 B8=1.07924826 B9=1.08212832 B10=1.08410104 B11=1.08448513 A1=125.42622743 A2=123.29927667 A3=113.32719559 A4=117.45192601 A5=121.85446325 A6=107.63531568 A7=107.58796065 A8=110.55833374 A9=110.16227437 A10=111.10365887 D1=-2.13060465 D2=-87.7293937 D3=-176.55919949 D4=179.34834596 D5=149.97218516 D6=34.25154375 D7=60.43270112 D8=-179.83435772 D9=-60.05872636 |
|
| Table 3. N-Ethylformamide. Atomic coordinates, rs/ropt. | |||||||
| a (Å) | b (Å) | c (Å) | |||||
| C | 1.281521 | 0.280872 | - | 0.252688 | |||
| O | 1.511462 | - | 0.835396 | 0.174971 | |||
| N | 0.078050 | 0.930079 | - | 0.101902 | |||
| C | - | 1.034828 | 0.297267 | 0.555192 | |||
| C | - | 1.817173 | - | 0.628812 | - | 0.354234 | |
| H | - | 0.037555 | 1.821794 | - | 0.559429 | ||
| H | 2.024607 | 0.889378 | - | 0.798052 | |||
| H | - | 1.680720 | 1.085157 | 0.954626 | |||
| H | - | 0.636495 | - | 0.264133 | 1.404619 | ||
| H | - | 1.167410 | - | 1.422596 | - | 0.730068 | |
| H | - | 2.646926 | - | 1.090075 | 0.190359 | ||
| H | - | 2.228511 | - | 0.080670 | - | 1.207163 | |
| Table 4. N-Ethylformamide. Rotational constants (MHz): rs/ropt and ropt = HF/aug-cc-pVTZ. | ||||
| rs/ropt | ropt | Expt. [1] | ||
| A | 9873.9 | 10590.3 |
9904.8373(6) | |
| B | 3555.3 | 3373.2 |
3521.0995(2) | |
| C | 3002.7 | 2935.8 |
2984.9808(2) | |