|
|
|
|
|
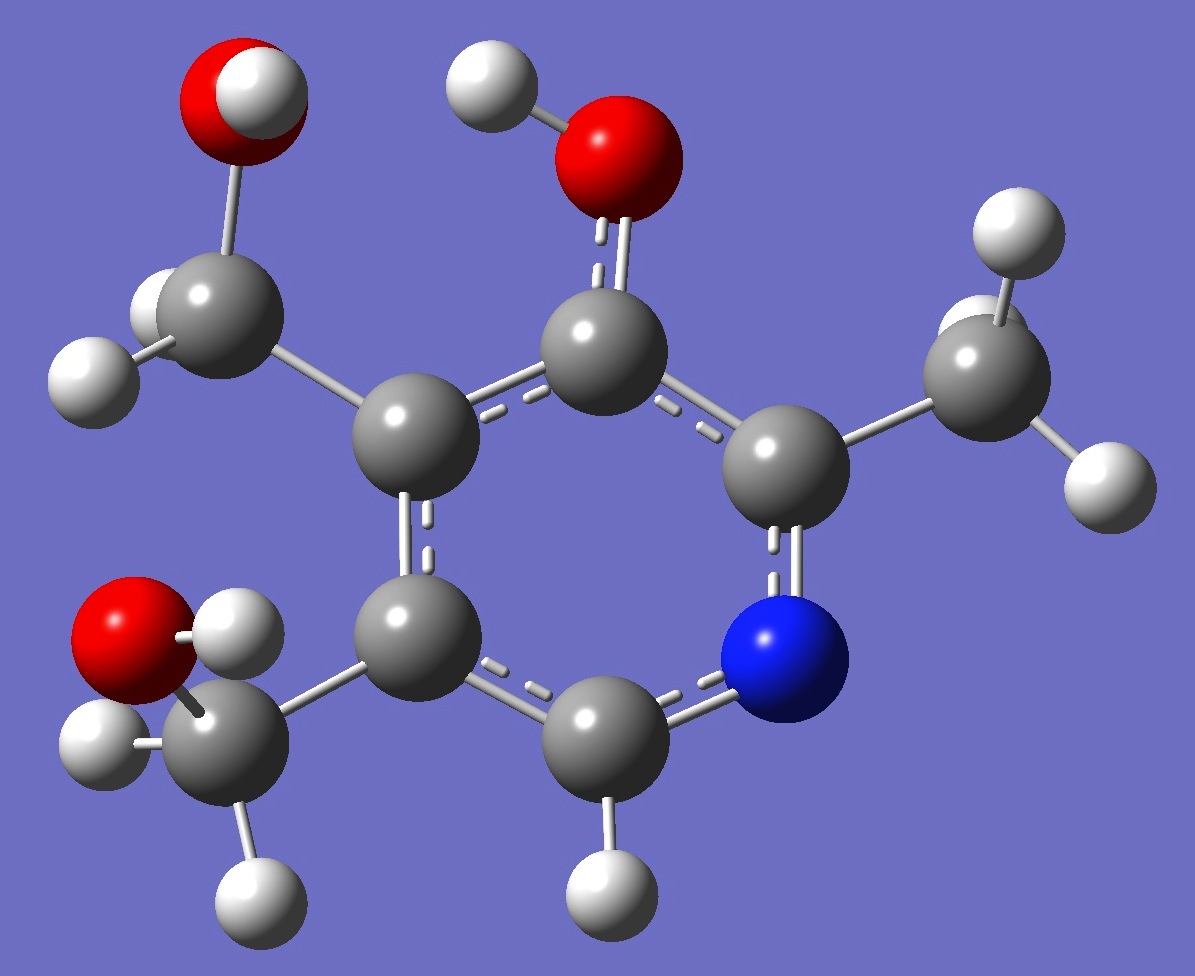
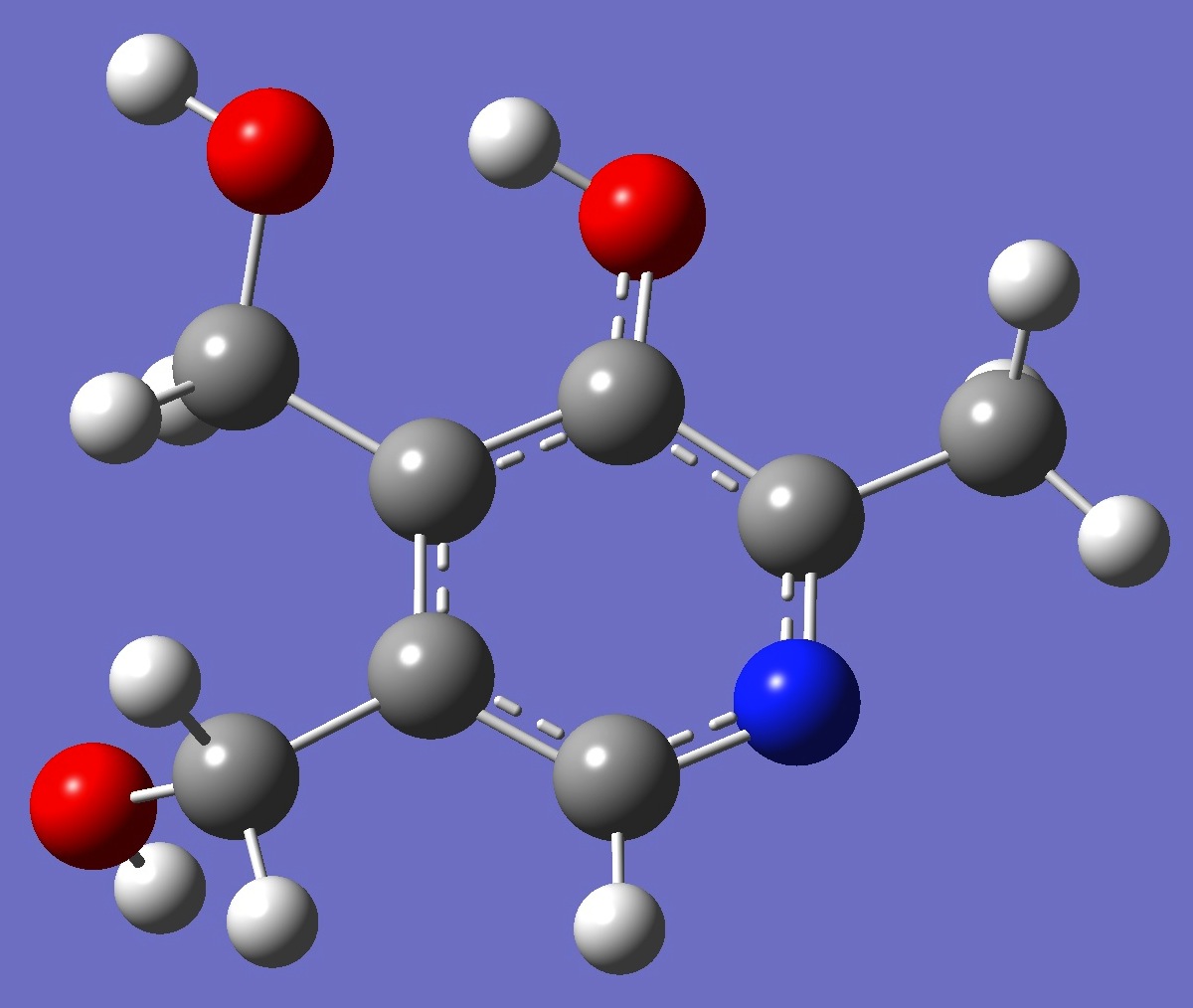
Table 3. Pyridoxine: B3P86/6-31G(3d,3p) optimized structure
parameters.
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
N,5,B5,4,A4,3,D3,0
H,1,B6,6,A5,5,D4,0
C,5,B7,4,A6,3,D5,0
H,8,B8,5,A7,4,D6,0
H,8,B9,5,A8,4,D7,0
H,8,B10,5,A9,4,D8,0
O,4,B11,3,A10,2,D9,0
H,12,B12,4,A11,3,D10,0
C,3,B13,2,A12,1,D11,0
H,14,B14,3,A13,2,D12,0
H,14,B15,3,A14,2,D13,0
C,2,B16,1,A15,6,D14,0
H,17,B17,2,A16,1,D15,0
H,17,B18,2,A17,1,D16,0
O,14,B19,3,A18,2,D17,0
H,20,B20,14,A19,3,D18,0
O,17,B21,2,A20,1,D19,0
H,22,B22,17,A21,2,D20,0
|
Conformer I |
|
|
|
|
|
|
|
|
B1=1.38862731
B2=1.40229365
B3=1.39483638
B4=1.40710975
B5=1.32933752
B6=1.08883704
B7=1.49566865
B8=1.09424768
B9=1.0938899
B10=1.08929508
B11=1.35234381
B12=0.97636483
B13=1.50674294
B14=1.09473849
B15=1.09108469
B16=1.50371356
B17=1.09361195
B18=1.09716719
B19=1.4329799
B20=0.96317413
B21=1.42270832
B22=0.96248292
A1=117.90719144
A2=117.73713501
A3=120.00180803
A4=121.50594664
A5=116.04778359
A6=119.86423437
A7=111.11309238
A8=111.06429916
A9=109.31778238
A10=122.46584064
|
A11=107.35535574
A12=122.58060799
A13=109.98961134
A14=110.97160259
A15=119.73669461
A16=111.63838898
A17=109.09776249
A18=112.58532253
A19=107.8517851
A20=112.89277066
A21=107.4811201
D1=-0.74442454
D2=1.09543736
D3=-0.59292798
D4=-179.4722054
D5=179.14931085
D6=-61.27829269
D7=56.90548009
D8=178.01687413
D9=-179.7610921
D10=16.73208308
D11=177.02194844
D12=-103.3744596
D13=16.55233092
D14=-179.21755672
D15=-125.87316467
D16=-7.97696989
D17=140.39908653
D18=-69.29214378
D19=115.82883838
D20=-57.27850384
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
N,5,B5,4,A4,3,D3,0
H,1,B6,6,A5,5,D4,0
C,5,B7,4,A6,3,D5,0
H,8,B8,5,A7,4,D6,0
H,8,B9,5,A8,4,D7,0
H,8,B10,5,A9,4,D8,0
O,4,B11,3,A10,2,D9,0
H,12,B12,4,A11,3,D10,0
C,3,B13,2,A12,1,D11,0
H,14,B14,3,A13,2,D12,0
H,14,B15,3,A14,2,D13,0
C,2,B16,1,A15,6,D14,0
H,17,B17,2,A16,1,D15,0
H,17,B18,2,A17,1,D16,0
O,14,B19,3,A18,2,D17,0
H,20,B20,14,A19,3,D18,0
O,17,B21,2,A20,1,D19,0
H,22,B22,17,A21,2,D20,0
|
Conformer II
|
|
|
|
|
|
|
|
|
B1=1.38800423
B2=1.40294327
B3=1.3955853
B4=1.40791668
B5=1.32881281
B6=1.08870803
B7=1.49582194
B8=1.09424592
B9=1.0938661
B10=1.08932195
B11=1.3516256
B12=0.97582679
B13=1.50315159
B14=1.09808753
B15=1.09307989
B16=1.50100534
B17=1.09711761
B18=1.09478511
B19=1.43277398
B20=0.961196
B21=1.42216487
B22=0.96260435
A1=118.18762906
A2=117.5625114
A3=119.82302487
A4=121.83629728
A5=116.14691775
A6=119.64433199
A7=111.16113061
A8=111.01288681
A9=109.26800366
A10=123.20290687
|
A11=107.12864766
A12=121.08863794
A13=109.47167028
A14=110.68019439
A15=119.88113982
A16=109.2475334
A17=111.18379102
A18=109.55201146
A19=108.53407876
A20=113.05362415
A21=107.52998328
D1=-0.67649778
D2=0.48912079
D3=-0.02520425
D4=-179.67784932
D5=179.51172819
D6=-60.65971065
D7=57.54289193
D8=178.56480226
D9=179.53343308
D10=14.76970211
D11=176.47456552
D12=-93.30515665
D13=24.47169463
D14=178.82207353
D15=5.31822772
D16=123.11880695
D17=145.86607822
D18=169.14642811
D19=-118.89630856
D20=57.85471889
|
|
|
|
|
|
|
|