|
|
|
|
|
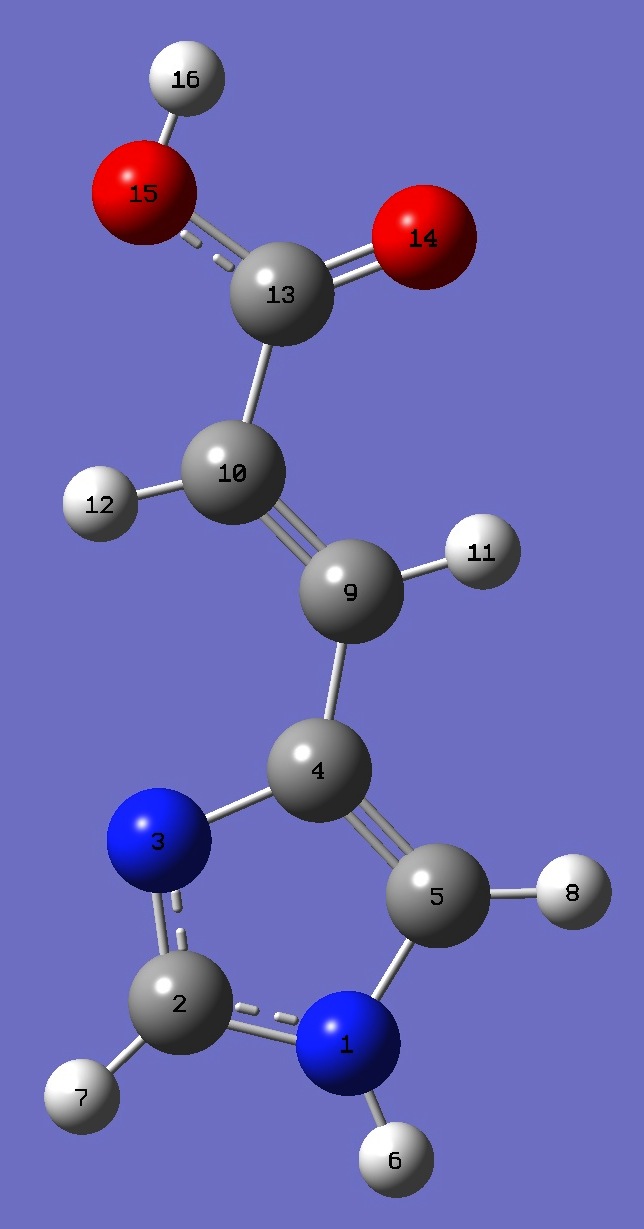
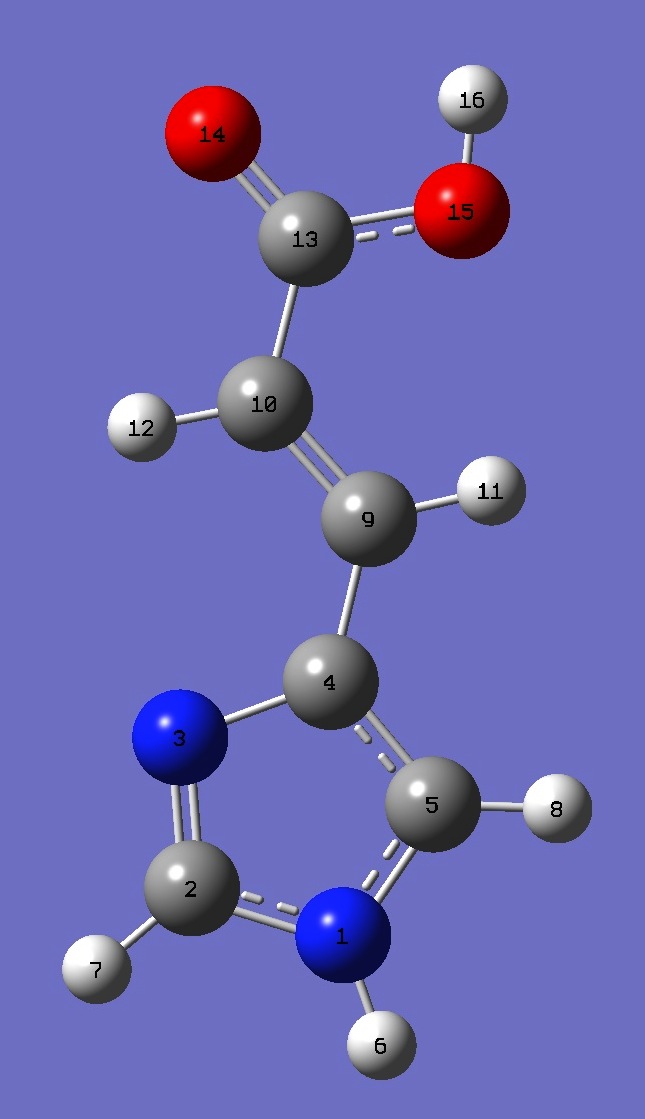
Table 4. Urocanic Acid: B3LYP/cc-pVTZ optimized structure
parameters.
|
|
|
|
|
|
N
C,1,B1
N,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
H,1,B5,2,A4,3,D3,0
H,2,B6,1,A5,5,D4,0
H,5,B7,4,A6,3,D5,0
C,4,B8,3,A7,2,D6,0
C,9,B9,4,A8,3,D7,0
H,9,B10,4,A9,3,D8,0
H,10,B11,9,A10,4,D9,0
C,10,B12,9,A11,4,D10,0
O,13,B13,10,A12,9,D11,0
O,13,B14,10,A13,9,D12,0
H,15,B15,13,A14,10,D13,0
|
Conformer A
|
|
|
|
|
|
|
|
|
|
B1=1.36712001
B2=1.30346416
B3=1.38361167
B4=1.37808517
B5=1.00516349
B6=1.07753796
B7=1.07509596
B8=1.44126525
B9=1.33911284
B10=1.08440362
B11=1.08002779
B12=1.46792393
B13=1.21056657
B14=1.35969503
B15=0.96786688
A1=111.71639053
A2=105.86432695
A3=109.5700965
A4=126.55083814
A5=122.27519923
A6=131.8234638
|
A7=123.65836848
A8=125.31086135
A9=116.84629359
A10=121.42056516
A11=120.31549462
A12=126.66567969
A13=111.48525771
A14=106.23861945
D1=0.
D2=0.
D3=180.
D4=180.
D5=180.
D6=180.
D7=0.
D8=180.
D9=0.
D10=180.
D11=0.
D12=180.
D13=180.
|
|
|
|
|
|
|
|
|
|
|
N
C,1,B1
N,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,1,B4,2,A3,3,D2,0
H,1,B5,2,A4,3,D3,0
H,2,B6,1,A5,5,D4,0
H,5,B7,1,A6,2,D5,0
C,4,B8,3,A7,2,D6,0
C,9,B9,4,A8,3,D7,0
H,9,B10,4,A9,3,D8,0
H,10,B11,9,A10,4,D9,0
C,10,B12,9,A11,4,D10,0
O,13,B13,10,A12,9,D11,0
O,13,B14,10,A13,9,D12,0
H,15,B15,13,A14,10,D13,0
|
Conformer B
|
|
|
|
|
|
|
|
|
|
B1=1.36732171
B2=1.30330029
B3=1.38332361
B4=1.36855711
B5=1.00516544
B6=1.07752516
B7=1.07513985
B8=1.44311659
B9=1.34013073
B10=1.08305632
B11=1.08004551
B12=1.46427579
B13=1.20961519
B14=1.36379324
B15=0.9672414
A1=111.70653533
A2=105.89463576
A3=107.1817397
A4=126.53232259
A5=122.25268894
A6=122.52706615
|
A7=123.58808802
A8=124.76569374
A9=116.23580272
A10=120.95662133
A11=124.01128287
A12=124.4590089
A13=114.0738991
A14=105.75531372
D1=0.
D2=0.
D3=180.
D4=180.
D5=180.
D6=180.
D7=0.
D8=180.
D9=0.
D10=180.
D11=180.
D12=0.
D13=180.
|
|
|
|
|
|
|
|
|
|
|
N
C,1,B1
N,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,1,B4,2,A3,3,D2,0
H,1,B5,2,A4,3,D3,0
H,2,B6,1,A5,5,D4,0
H,5,B7,1,A6,2,D5,0
C,4,B8,3,A7,2,D6,0
C,9,B9,4,A8,3,D7,0
H,9,B10,4,A9,3,D8,0
H,10,B11,9,A10,4,D9,0
C,10,B12,9,A11,4,D10,0
O,13,B13,10,A12,9,D11,0
O,13,B14,10,A13,9,D12,0
H,15,B15,13,A14,10,D13,0
|
Conformer C
|
|
|

|
|
|
|
|
|
|
B1=1.36676185
B2=1.30449759
B3=1.38357134
B4=1.3664008
B5=1.00555512
B6=1.07719661
B7=1.07468369
B8=1.44377141
B9=1.33984683
B10=1.08367369
B11=1.08140122
B12=1.46792549
B13=1.20894308
B14=1.36180567
B15=0.96784874
A1=111.41221223
A2=106.09988868
A3=107.48151243
A4=126.46979391
A5=122.37886731
A6=122.10469703
|
A7=120.37737257
A8=126.955161
A9=115.0243855
A10=122.65593782
A11=120.6970715
A12=126.83826982
A13=111.23388097
A14=106.1718662
D1=0.
D2=0.
D3=180.
D4=180.
D5=180.
D6=180.
D7=180.
D8=0.
D9=0.
D10=180.
D11=0.
D12=180.
D13=180.
|
|
|
|
|
|
|
|


