| |
||||||||
| Table 1. 14N nqcc's in gauche 3 Allylamine (MHz). Calculation was made on (1) MP2/6-311+G(d,p) and (2) MP2/6-311+G(3df,3pd) optimized molecular structures. | ||||||||
| |
||||||||
| Calc (1) | Calc (2) | Expt [1] |
||||||
| |
||||||||
| Xaa | 2.421 |
2.414 |
2.31(13) |
|||||
| Xbb | - |
0.078 |
- |
0.054 |
- |
0.51(8) |
||
| Xcc | - |
2.343 |
- |
2.360 |
- |
1.80(8) |
||
| Xab | 0.044 |
0.049 |
||||||
| Xac | 0.694 |
0.692 |
||||||
| Xbc | 3.337 |
3.320 |
||||||
| RMS |
0.406 (26. %) |
0.422 (27. %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.833 |
2.828 |
||||||
| Xyy | 1.942 | 1.934 | ||||||
| Xzz | - |
4.775 |
- |
4.762 |
||||
| ETA | - |
0.187 |
- |
0.187 |
||||
| |
||||||||
| Table 2. 14N nqcc's in gauche 2 Allylamine (MHz). Calculation was made on (1) MP2/6-311+G(d,p) and (2) MP2/6-311+G(3df,3pd) optimized molecular structures. | ||||||||
| |
||||||||
| Calc (1) | Calc (2) | Expt [2] |
||||||
| |
||||||||
| Xaa | - |
1.522 |
- |
1.646 |
- |
1.48(4) |
||
| Xbb | - |
0.019 |
0.300 |
0.03(3) |
||||
| Xcc | 1.540 |
1.346 |
1.45(3) |
|||||
| Xab | 3.454 |
3.305 |
||||||
| Xac | - |
1.124 |
- |
1.401 |
||||
| Xbc | 1.136 |
1.199 |
||||||
| RMS |
0.064 (6.5 %) |
0.193 (20. %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.800 |
2.788 |
||||||
| Xyy | 1.908 | 1.892 | ||||||
| Xzz | - |
4.709 |
- |
4.680 |
||||
| ETA | - |
0.189 |
- |
0.191 |
||||
| |
||||||||
| Table 3. 14N nqcc's in gauche 1 Allylamine (MHz). Calculation was made on (1) MP2/6-311+G(d,p) and (2) MP2/6-311+G(3df,3pd) optimized molecular structures. | ||||||||
| |
||||||||
| Calc (1) | Calc (2) | Expt |
||||||
| |
||||||||
| Xaa | 2.135 |
|||||||
| Xbb | 1.766 |
|||||||
| Xcc | - |
3.901 |
||||||
| Xab | 0.794 |
|||||||
| Xac | 1.746 |
|||||||
| Xbc | - |
1.521 |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xxx | 2.814 |
|||||||
| Xyy | 1.984 | |||||||
| Xzz | - |
4.798 |
||||||
| ETA | - |
0.173 |
||||||
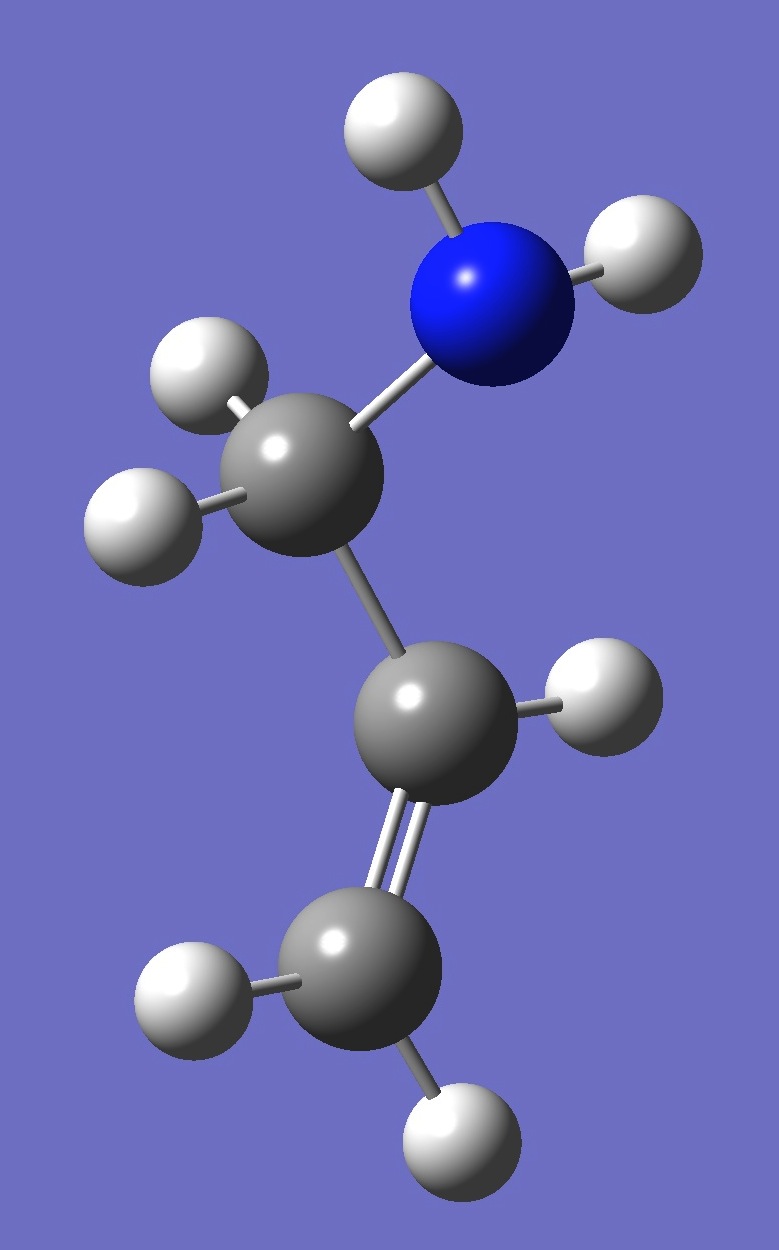
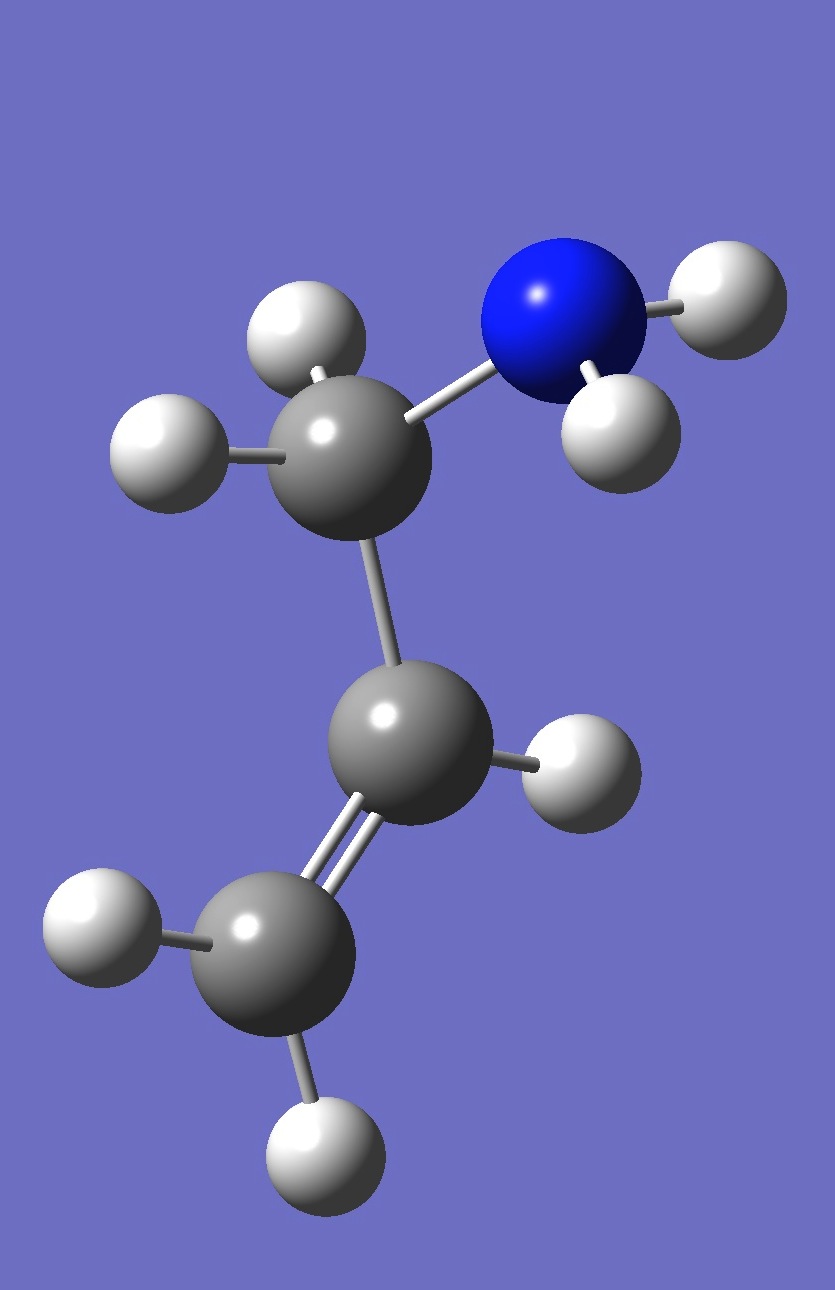
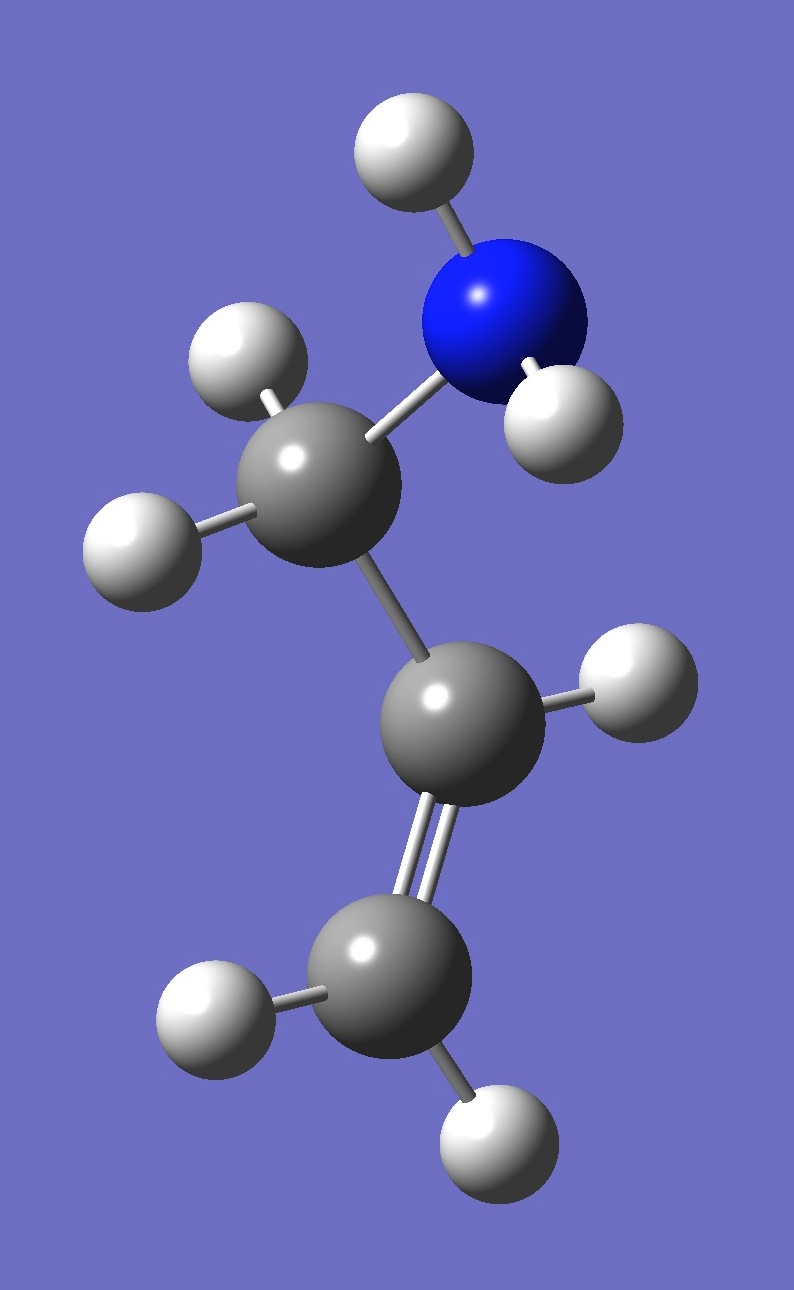
| Table 4. gauche Allylamine. Optimized structure parameters
(Å
and degrees). MP2/6-311+G(d,p) and
MP2/6-311+G(3df,3pd). |
||
|
C C,1,B1 H,1,B2,2,A1 H,2,B3,1,A2,3,D1,0 H,2,B4,1,A3,3,D2,0 C,1,B5,2,A4,4,D3,0 H,6,B6,1,A5,2,D4,0 H,6,B7,1,A6,2,D5,0 N,6,B8,1,A7,2,D6,0 H,9,B9,6,A8,1,D7,0 H,9,B10,6,A9,1,D8,0 |
||
| |
||
| gauche 3 |
||
| |
||
| MP2/6-311+G(d,p) |
MP2/6-311+G(3df,3pd) |
|
| |
||
| B1=1.34137412 B2=1.08893911 B3=1.08518642 B4=1.08717587 B5=1.50085873 B6=1.10088901 B7=1.09615554 B8=1.46877734 B9=1.01545815 B10=1.0157099 A1=120.31895604 A2=121.50333734 A3=121.07990468 A4=123.63919596 A5=108.9744303 A6=109.37842244 A7=109.44404372 A8=110.51825196 A9=109.64363645 D1=-1.19049515 D2=179.18217647 D3=-179.54972479 D4=-3.56174638 D5=-120.77592808 D6=121.86418423 D7=176.29741037 D8=-65.77048804 |
B1=1.33424501 B2=1.0843469 B3=1.08051186 B4=1.08237587 B5=1.49474292 B6=1.09565463 B7=1.09138695 B8=1.46507831 B9=1.01244154 B10=1.01288207 A1=120.13636867 A2=121.52096544 A3=120.92366034 A4=123.79411365 A5=109.12785536 A6=109.39900386 A7=109.39590087 A8=110.64826031 A9=109.74410415 D1=-1.19788358 D2=179.135458 D3=-179.61838338 D4=-3.519728 D5=-120.83665681 D6=121.98189046 D7=176.45404342 D8=-65.75369968 |
|
| gauche 2 |
||
| B1=1.3420617 B2=1.09106636 B3=1.08529541 B4=1.08730153 B5=1.50564773 B6=1.09525594 B7=1.09562231 B8=1.46915987 B9=1.01595901 B10=1.01561809 A1=119.56823208 A2=121.71168295 A3=120.89321793 A4=123.85404176 A5=109.2332093 A6=110.04313016 A7=114.71347004 A8=109.53970752 A9=110.32928748 D1=-0.55467656 D2=179.74077571 D3=-178.64147419 D4=-4.35405545 D5=-121.58742709 D6=117.20292331 D7=-52.06024954 D8=64.99180436 |
B1=1.33496638 B2=1.08640922 B3=1.08066962 B4=1.08264117 B5=1.49892757 B6=1.09055062 B7=1.09072777 B8=1.465917 B9=1.01336172 B10=1.01305861 A1=119.40002644 A2=121.70912386 A3=120.76627888 A4=124.01981577 A5=109.3757399 A6=110.20487674 A7=114.35961328 A8=109.40013673 A9=110.1280672 D1=-0.56486308 D2=179.74742237 D3=-178.40174775 D4=-5.7166826 D5=-123.13409912 D6=115.82285843 D7=-52.46837331 D8=63.92663859 |
|
| gauche 1 |
||
| C C,1,B1 H,1,B2,2,A1 H,2,B3,1,A2,3,D1,0 H,2,B4,1,A3,3,D2,0 C,1,B5,2,A4,4,D3,0 H,6,B6,1,A5,2,D4,0 H,6,B7,1,A6,2,D5,0 N,6,B8,1,A7,2,D6,0 H,9,B9,6,A8,1,D7,0 H,9,B10,6,A9,1,D8,0 Variables: B1=1.34021292 B2=1.09086996 B3=1.08511419 B4=1.08667561 B5=1.50114157 B6=1.09452236 B7=1.10221701 B8=1.47162007 B9=1.01627085 B10=1.01618726 A1=119.42247925 A2=121.44714806 A3=121.07929199 A4=124.21529745 A5=109.24150071 A6=108.67556777 A7=110.21403878 A8=109.69089471 A9=109.93396467 D1=-0.0675607 D2=179.75097017 D3=179.2690471 D4=3.11659472 D5=-113.8914931 D6=121.81298945 D7=59.96633896 D8=176.79515358 |
||