|
| |
|
|
|
|
|
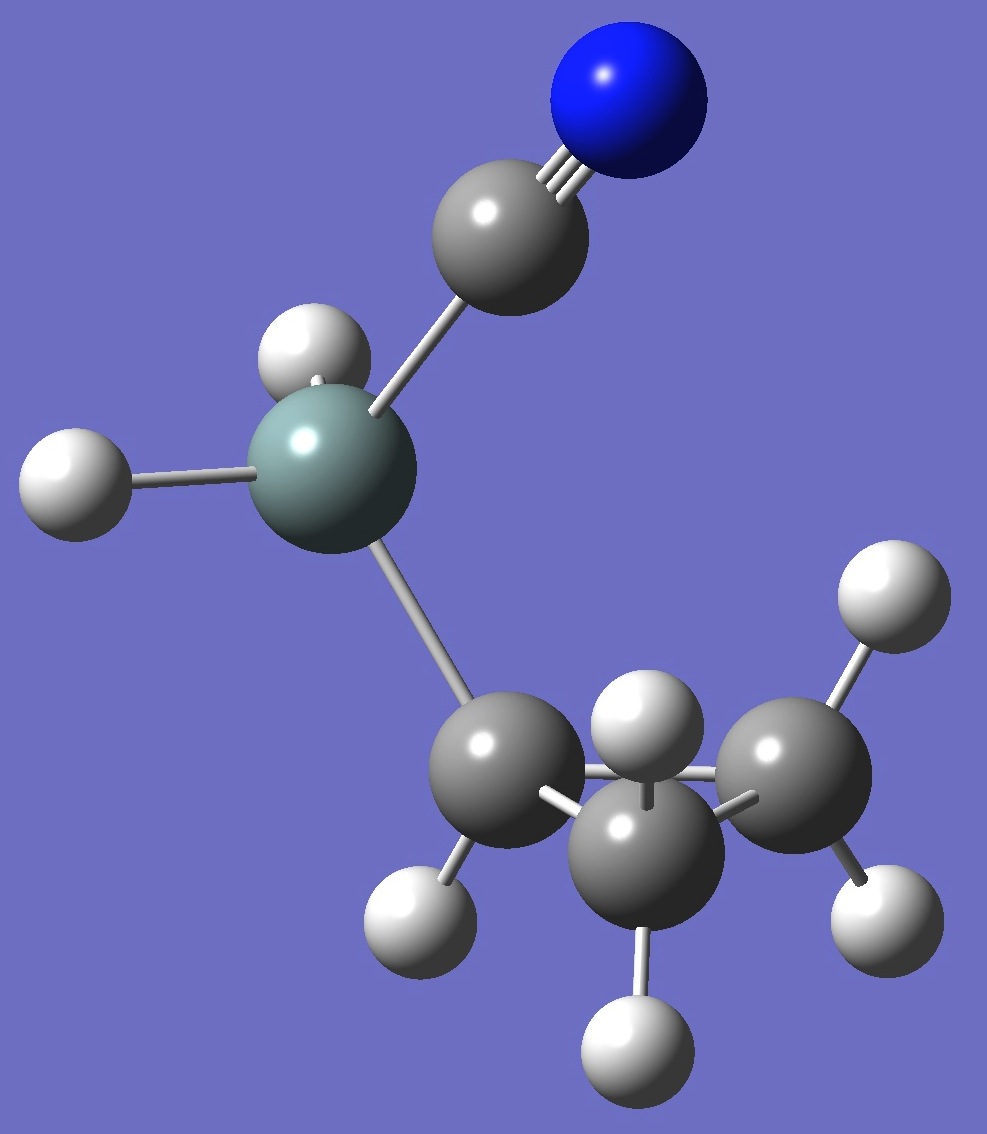
| Table 3. Cyclopropyl Cyanosilane. B3P86/6-31G(3d,3p) and mPW1PW91/6-31G(3d,3p) optimized structure parameters
(Å and degrees). |
| |
|
|
|
|
|
|
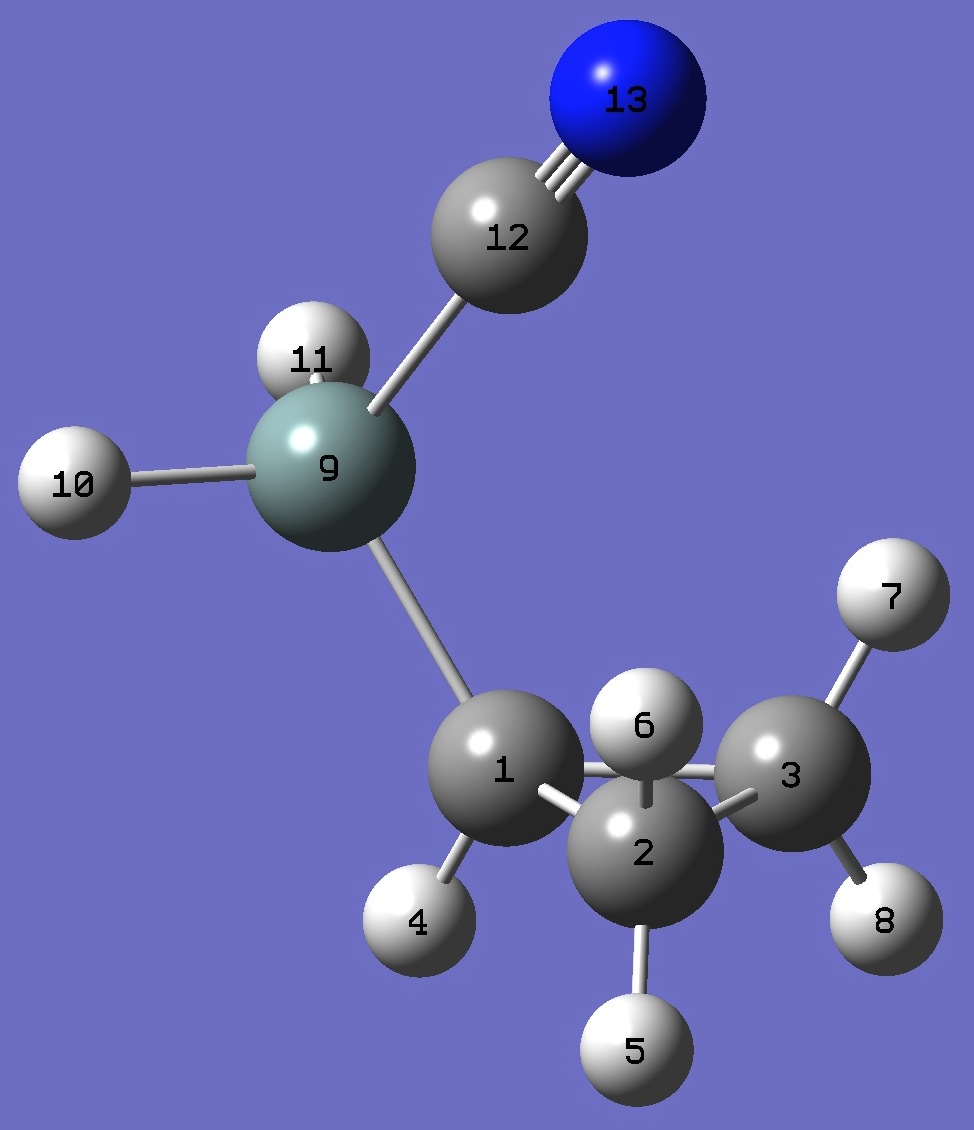
cis Cyclopropyl Cyanosilane
|
|
|
|
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
H,1,B3,2,A2,3,D1,0
H,2,B4,1,A3,3,D2,0
H,2,B5,1,A4,3,D3,0
H,3,B6,2,A5,1,D4,0
H,3,B7,2,A6,1,D5,0
Si,1,B8,2,A7,3,D6,0
H,9,B9,1,A8,2,D7,0
H,9,B10,1,A9,2,D8,0
C,9,B11,1,A10,2,D9,0
N,12,B12,9,A11,1,D10,0
|
|
|
|
|
|
|
|
|
|
B3P86
|
mPW1PW91
|
|
|
|
|
|
|
|
|
|
B1=1.51434544
B2=1.49039794
B3=1.08773916
B4=1.0835398
B5=1.08472477
B6=1.08472477
B7=1.0835398
B8=1.84229726
B9=1.47816639
B10=1.47816639
B11=1.85166783
B12=1.15863092
A1=60.52175075
A2=114.91007488
A3=117.06383777
A4=118.32616304
A5=117.71852488
A6=118.50968256
A7=121.39922998
A8=113.36320678
A9=113.36320678
A10=107.10486114
A11=177.4074877
D1=105.21912214
D2=-109.08028992
D3=107.59431447
D4=108.5843395
D5=-106.72181861
D6=-110.18380538
D7=-81.91725313
D8=152.32954589
D9=35.20614638
D10=0.
|
B1=1.51357344
B2=1.48974208
B3=1.08705639
B4=1.0829037
B5=1.08402418
B6=1.08402418
B7=1.0829037
B8=1.84355909
B9=1.47913251
B10=1.47913251
B11=1.85368019
B12=1.15696855
A1=60.51949114
A2=114.92923994
A3=117.02116153
A4=118.34669295
A5=117.74285526
A6=118.47763125
A7=121.4305515
A8=113.34336879
A9=113.34336879
A10=107.20887021
A11=177.42628096
D1=105.23420962
D2=-109.05906622
D3=107.61426609
D4=108.59804886
D5=-106.68274794
D6=-110.21164949
D7=-81.91453071
D8=152.35946954
D9=35.22246941
D10=0.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
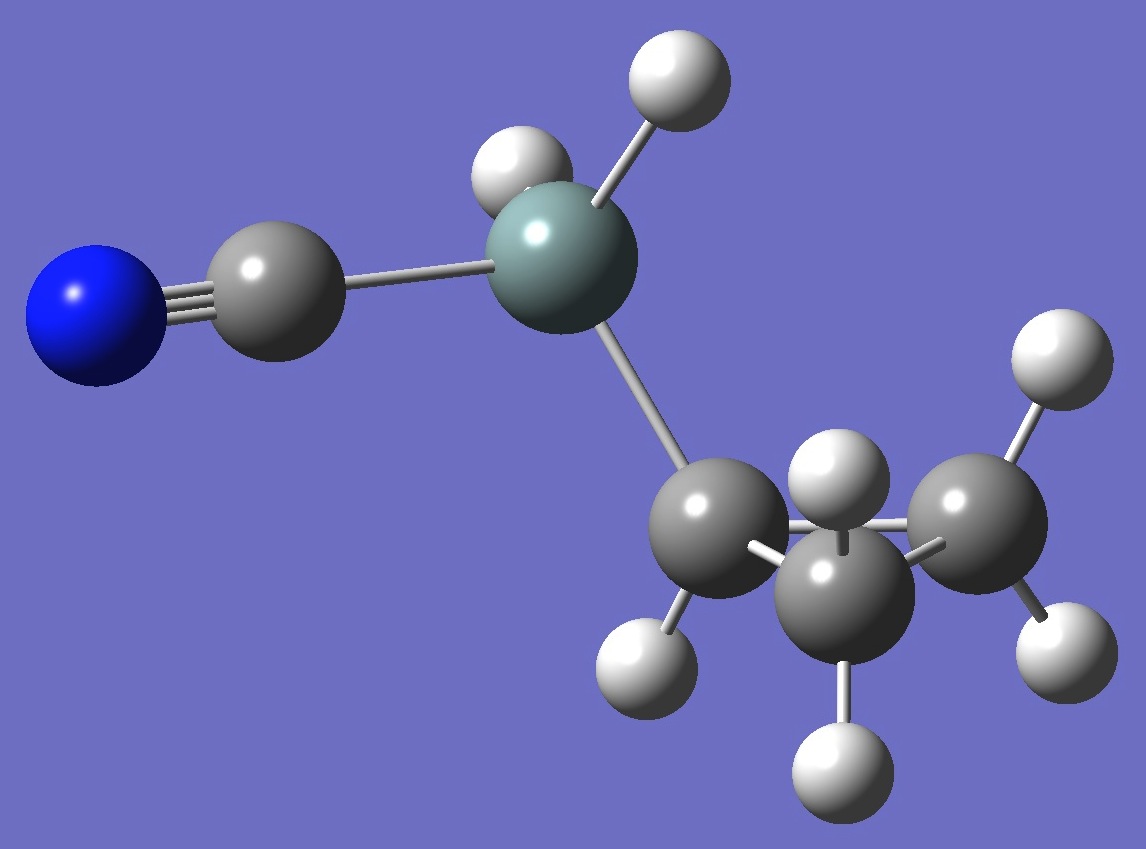
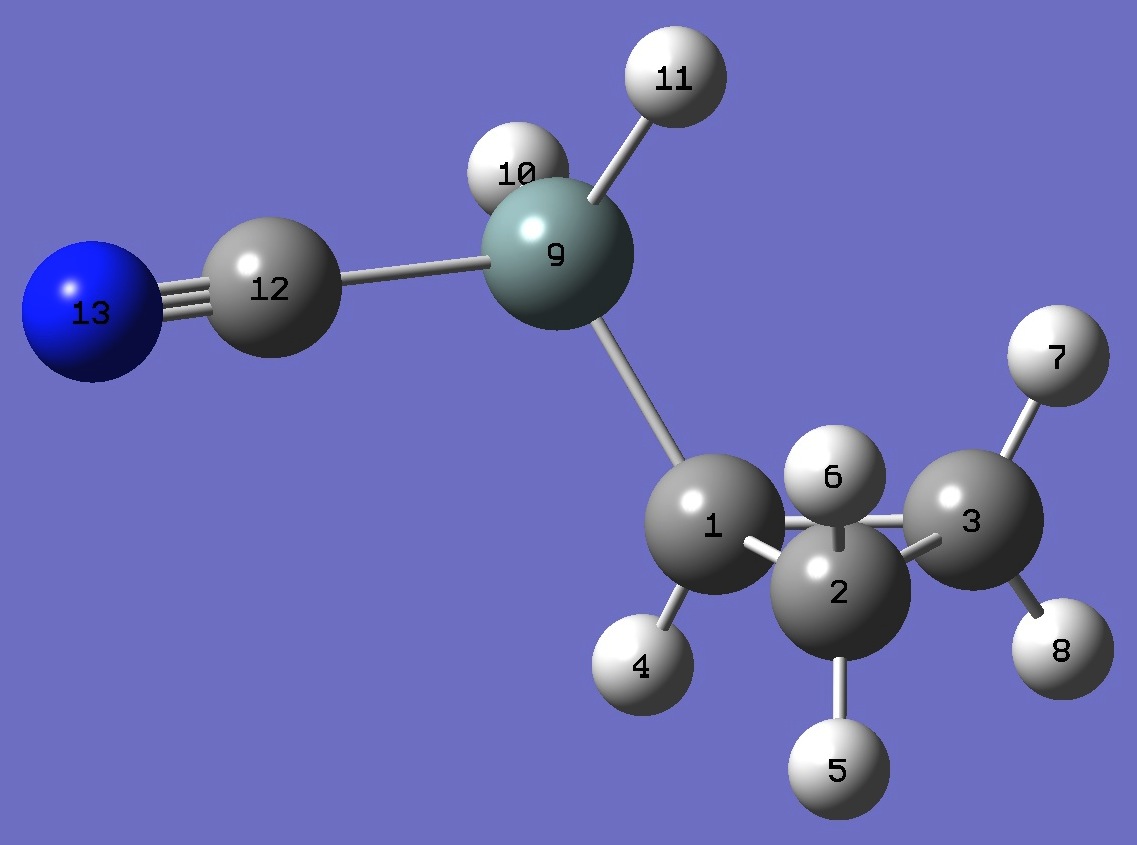
gauche Cyclopropyl Cyanosilane |
|
|
|
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
H,1,B3,2,A2,3,D1,0
H,2,B4,1,A3,3,D2,0
H,2,B5,1,A4,3,D3,0
H,3,B6,2,A5,1,D4,0
H,3,B7,2,A6,1,D5,0
Si,1,B8,2,A7,3,D6,0
H,9,B9,1,A8,2,D7,0
H,9,B10,1,A9,2,D8,0
C,9,B11,1,A10,2,D9,0
N,12,B12,1,A11,9,D10,0
|
|
|
|
|
|
|
|
|
|
B3P86
|
mPW1PW91
|
|
|
|
|
|
|
|
|
|
B1=1.51338575
B2=1.48993982
B3=1.08657386
B4=1.08341275
B5=1.08472588
B6=1.08498375
B7=1.08343902
B8=1.83931679
B9=1.477847
B10=1.47938242
B11=1.85113806
B12=1.15847136
A1=60.67019268
A2=115.16291264
A3=116.98178696
A4=118.42355626
A5=118.12862134
A6=118.52059465
A7=120.37025103
A8=113.4310095
A9=110.17629106
A10=110.27751387
A11=144.34788552
D1=105.25087772
D2=-109.05863285
D3=108.21014283
D4=108.57495764
D5=-106.64598969
D6=-108.05454463
D7=158.50281202
D8=34.19464919
D9=-83.0734243
D10=179.52741074
|
B1=1.51277557
B2=1.48927353
B3=1.08587952
B4=1.08273138
B5=1.08402543
B6=1.08427856
B7=1.08276785
B8=1.84057018
B9=1.47864022
B10=1.4801979
B11=1.85279531
B12=1.15682708
A1=60.664694
A2=115.16704634
A3=116.96539204
A4=118.41443378
A5=118.12242264
A6=118.49611676
A7=120.35134591
A8=113.40098323
A9=110.24473908
A10=110.28767458
A11=144.39698396
D1=105.28299302
D2=-109.03255348
D3=108.201219
D4=108.57846189
D5=-106.62393831
D6=-108.06910021
D7=158.48475601
D8=34.18938953
D9=-83.11769605
D10=179.53508545
|
|
|
|
|
|
|
|
|
|