|
| |
|
|
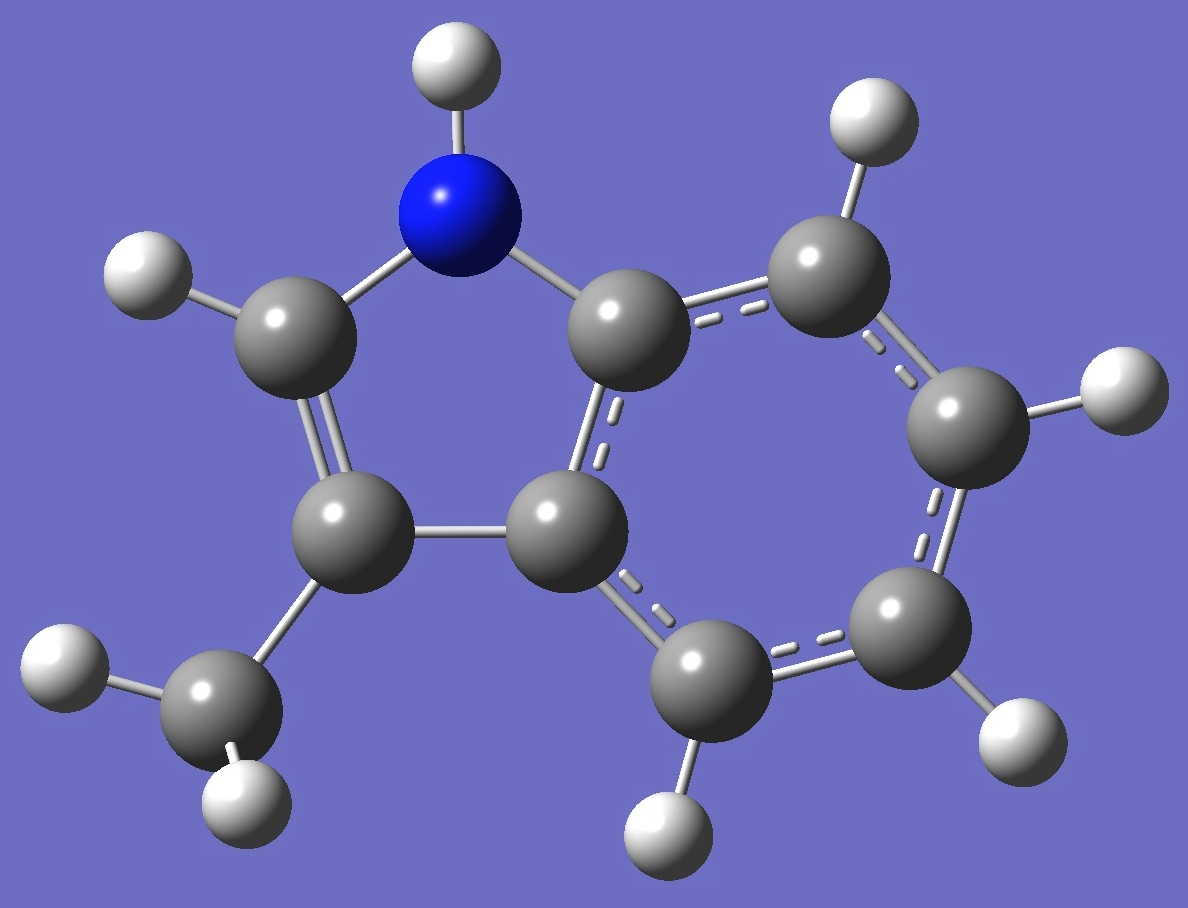
| Table 2. 3-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
|
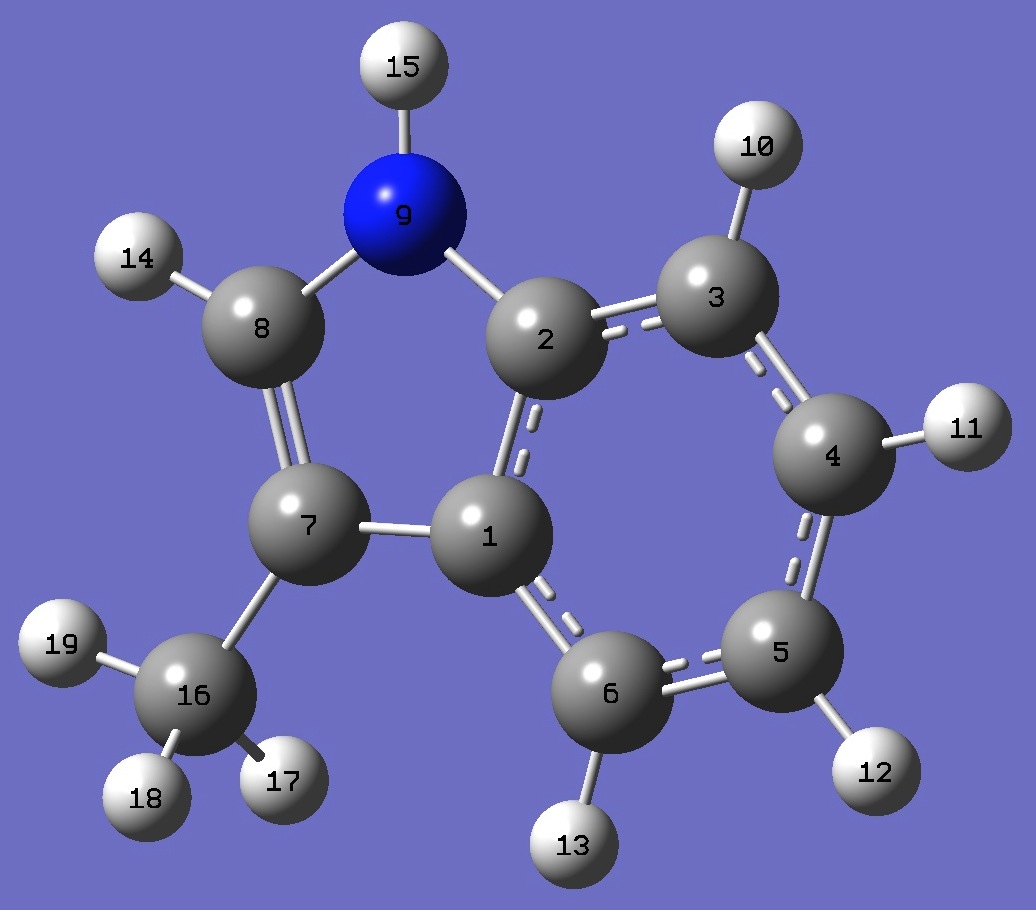
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,4,B10,3,A9,2,D8,0
H,5,B11,4,A10,3,D9,0
H,6,B12,5,A11,4,D10,0
H,8,B13,7,A12,1,D11,0
H,9,B14,2,A13,1,D12,0
C,7,B15,1,A14,6,D13,0
H,16,B16,7,A15,1,D14,0
H,16,B17,7,A16,1,D15,0
H,16,B18,7,A17,1,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41797381
B2=1.39347
B3=1.38461831
B4=1.40466455
B5=1.38360577
B6=1.43858546
B7=1.36635269
B8=1.37434695
B9=1.08245761
B10=1.08178196
B11=1.08178353
B12=1.08239568
B13=1.07736021
B14=1.00275712
B15=1.49403168
B16=1.09307719
B17=1.09307719
B18=1.09020921
A1=122.19748779
A2=117.59668951
A3=121.19837608
A4=121.05960069
A5=133.77512412
A6=106.22979334
A7=107.08824279
A8=121.39377675
A9=119.41555133
A10=119.20154155
A11=120.34437003
A12=129.34964762
A13=125.60886727
A14=126.74809078
A15=111.62264588
A16=111.62264588
A17=111.1827846
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=180.
D12=180.
D13=0.
D14=-59.80899315
D15=59.80899315
D16=180.
|
B1=1.4170632
B2=1.3933325
B3=1.38470127
B4=1.40442432
B5=1.38368698
B6=1.43664941
B7=1.36779974
B8=1.37263626
B9=1.08532397
B10=1.08486163
B11=1.08485379
B12=1.08556994
B13=1.08046592
B14=1.00464856
B15=1.49052117
B16=1.09529338
B17=1.09529338
B18=1.09240981
A1=122.3090373
A2=117.44599139
A3=121.26863265
A4=121.09040539
A5=133.73087321
A6=106.16239879
A7=107.0820375
A8=121.4712814
A9=119.36327504
A10=119.18815854
A11=120.46019636
A12=129.45250103
A13=125.48979947
A14=126.80655045
A15=111.63132069
A16=111.63132069
A17=111.16862946
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=180.
D12=180.
D13=0.
D14=-59.82319631
D15=59.82319631
D16=180.
|
|
|
|
|