|
| |
|
|
|
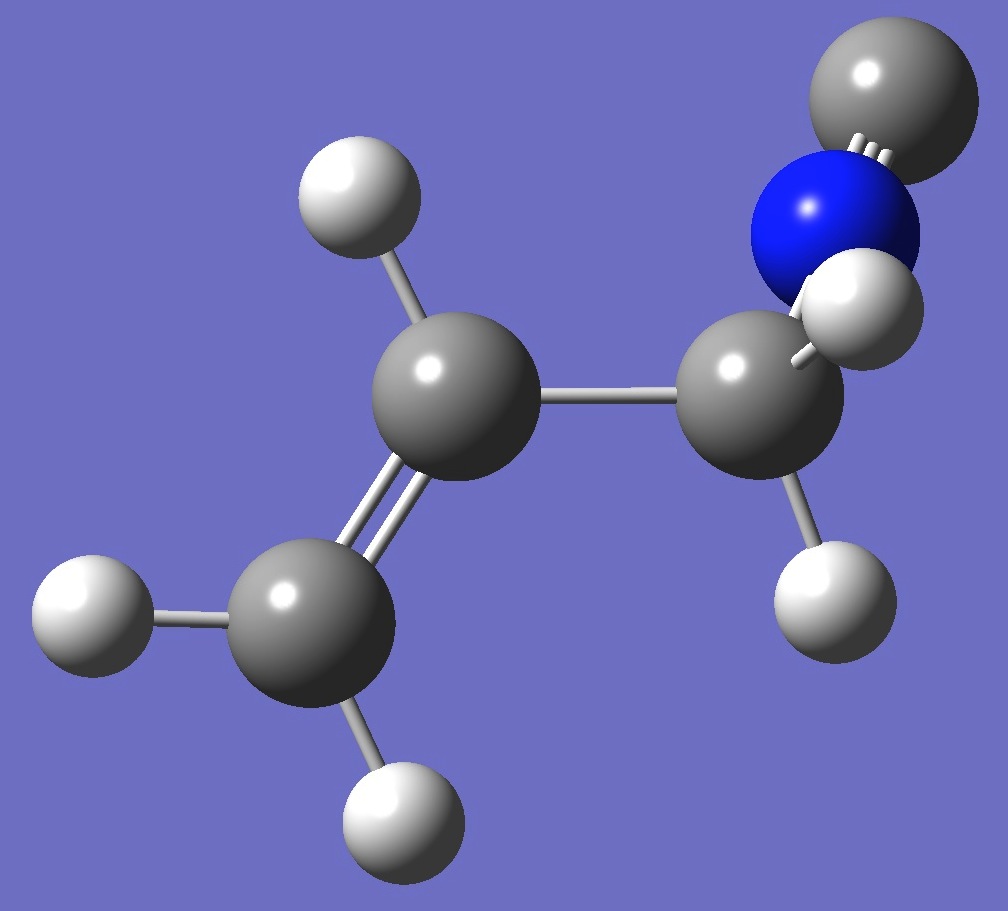
| Table 2. Molecular
structure parameters: ropt(1) = B3P86/6-31G(3d,3p), ropt(2) = B3P86/6-311+G(3df,3pd) (Å
and degrees). |
| |
|
|
|
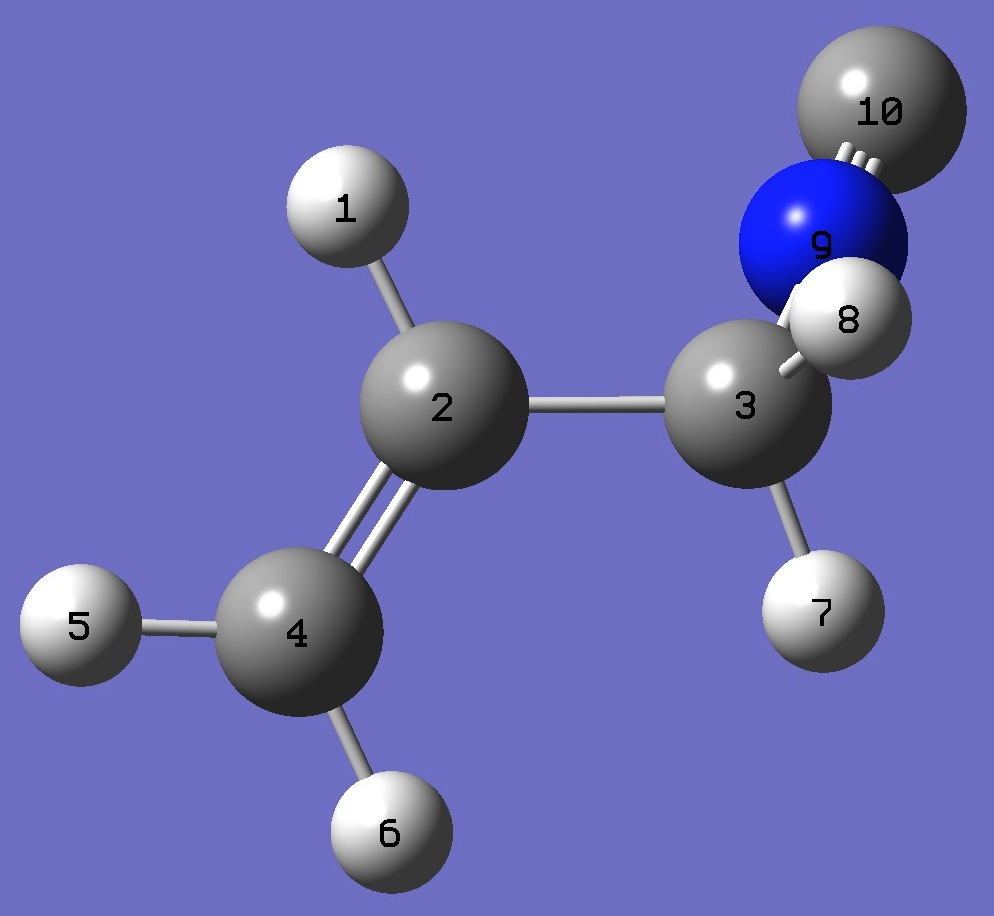
|
H
C,1,B1
C,2,B2,1,A1
C,2,B3,1,A2,3,D1,0
H,4,B4,2,A3,1,D2,0
H,4,B5,2,A4,1,D3,0
H,3,B6,2,A5,1,D4,0
H,3,B7,2,A6,1,D5,0
N,3,B8,2,A7,1,D6,0
C,9,B9,3,A8,2,D7,0
|
|
|
|
|
|
|
|
|
|
|
|
|
ropt(1) |
ropt(2) |
|
|
|
|
|
|
B1=1.08699486
B2=1.50075286
B3=1.32610949
B4=1.0843335
B5=1.08635791
B6=1.09372501
B7=1.09633211
B8=1.42299612
B9=1.17098417
A1=115.81074413
A2=120.88999085
A3=121.42197498
A4=121.65500893
A5=110.60759314
A6=110.52108073
A7=111.82892762
A8=178.51557809
D1=179.29215345
D2=-0.90243336
D3=179.32673426
D4=-176.50749599
D5=64.46577983
D6=-55.26977769
D7=19.57418831
|
B1=1.08433834
B2=1.49800615
B3=1.32204174
B4=1.08143588
B5=1.08353147
B6=1.0905446
B7=1.09322127
B8=1.41943641
B9=1.16512449
A1=115.79268825
A2=120.78621219
A3=121.36809655
A4=121.66347714
A5=110.54458754
A6=110.2919086
A7=111.98931007
A8=178.86445413
D1=179.03476802
D2=-0.80207237
D3=179.27551329
D4=-175.19557421
D5=65.91059516
D6=-53.79870768
D7=21.49306808
|
|
|
|
|
|
|
|
|
[1] I.Haykal, L.Margulès,
T.R.Huet, R.A.Motyienko, P.Écija, E.J.Cocinero, F.Basterretxea,
J.A.Fernández, F.Castaño, A.Lesarri, J.C.Guillemin, B.Tercero, and
J.Cernicharo, Ap.J. 777,120(2013).
|
|