|
|
|
|
|
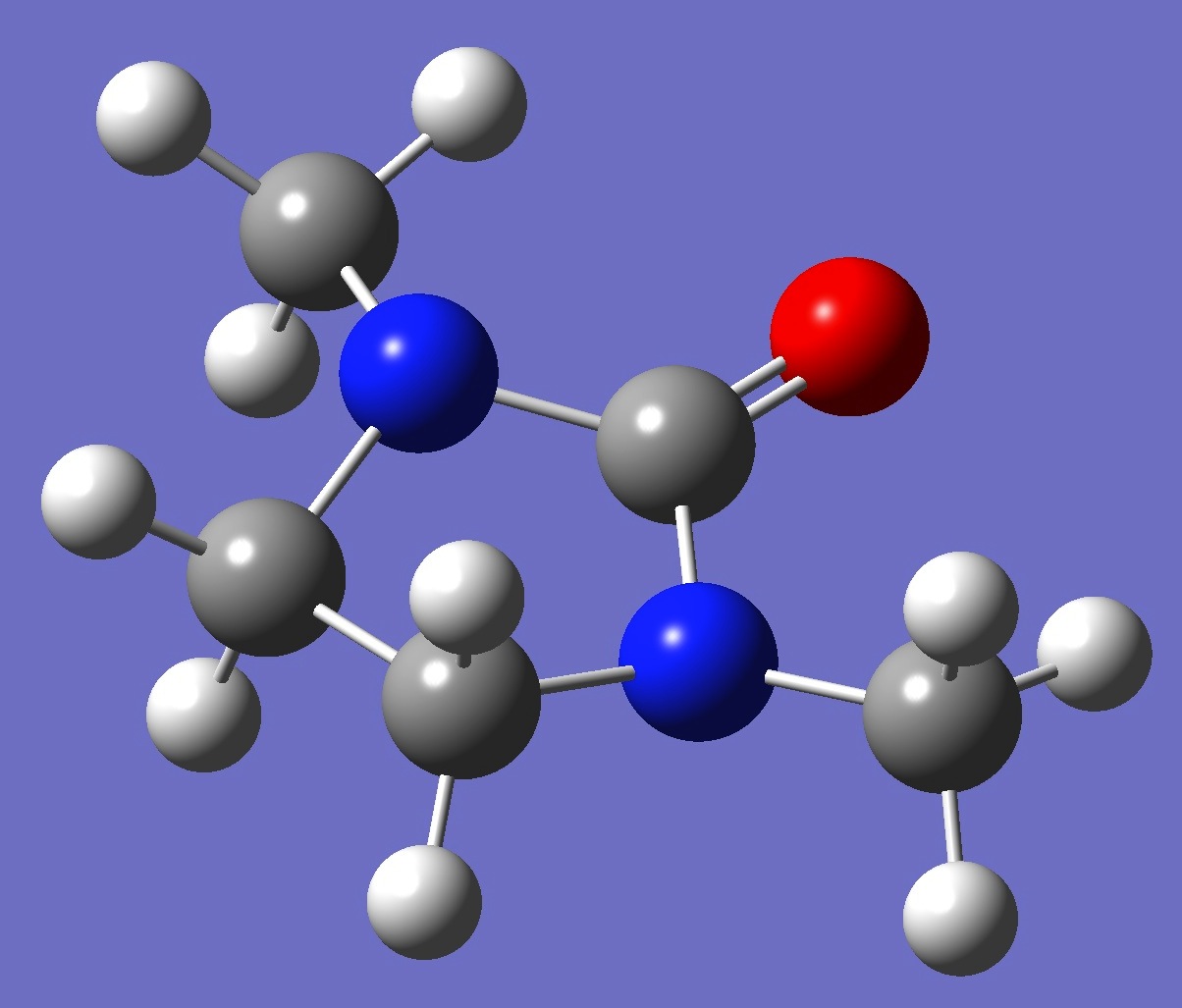
Table 2. 1,3-Dimethyl-2-imidazolidione: Structure parameters, ropt(1) and ropt(2) (Å and degrees).
|
|
|
|
|
|
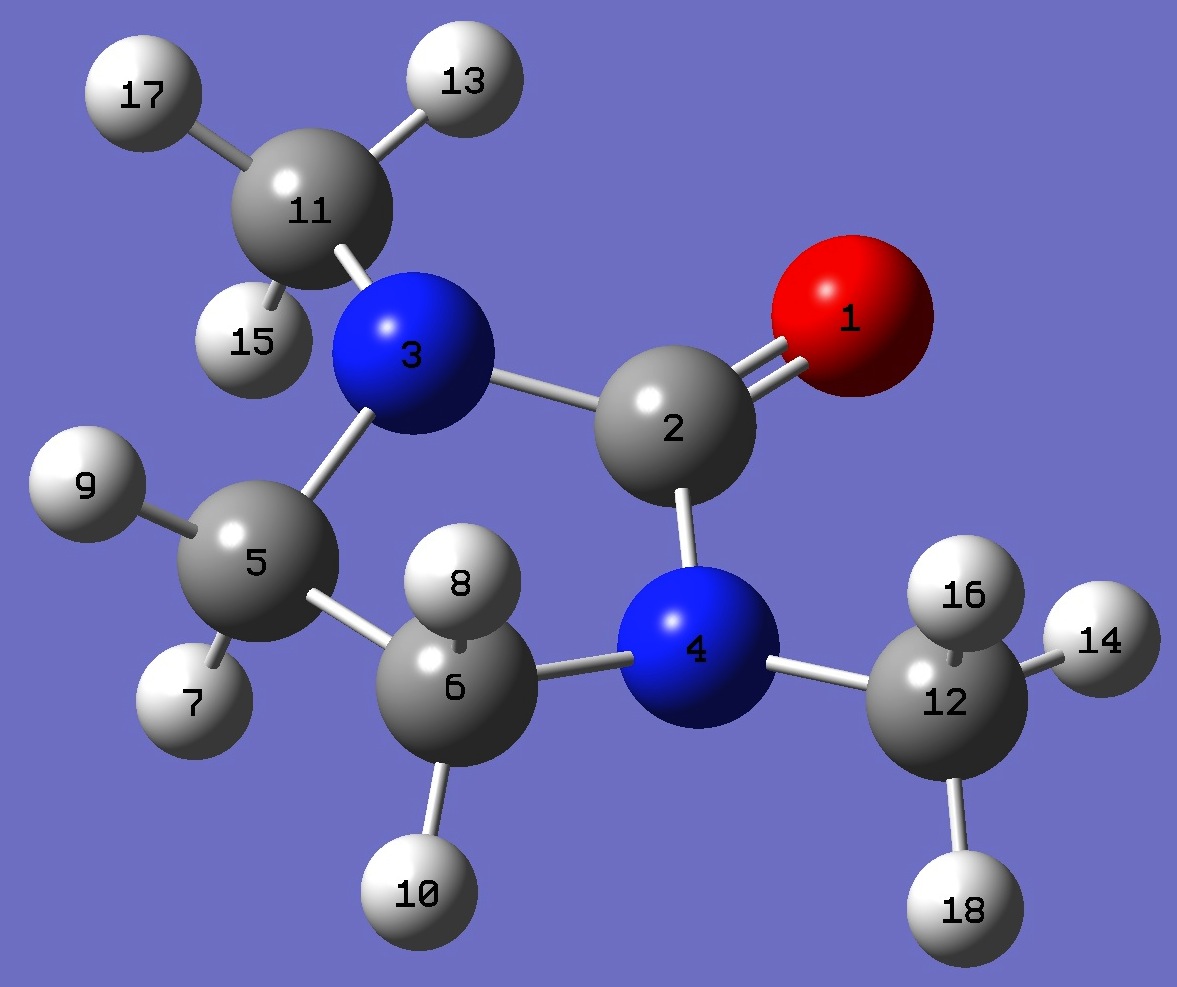
O
C,1,B1
N,2,B2,1,A1
N,2,B3,1,A2,3,D1,0
C,3,B4,2,A3,1,D2,0
C,4,B5,2,A4,1,D3,0
H,5,B6,3,A5,2,D4,0
H,6,B7,4,A6,2,D5,0
H,5,B8,3,A7,2,D6,0
H,6,B9,4,A8,2,D7,0
C,3,B10,2,A9,1,D8,0
C,4,B11,2,A10,1,D9,0
H,11,B12,3,A11,2,D10,0
H,12,B13,4,A12,2,D11,0
H,11,B14,3,A13,2,D12,0
H,12,B15,4,A14,2,D13,0
H,11,B16,3,A15,2,D14,0
H,12,B17,4,A16,2,D15,0
|
|
|
| ropt(1) |
ropt(2)
|
|
|
|
|
B1=1.22080009
B2=1.39310133
B3=1.39310133
B4=1.45411835
B5=1.45411835
B6=1.10207972
B7=1.10207972
B8=1.09302508
B9=1.09302508
B10=1.45028672
B11=1.45028672
B12=1.09004367
B13=1.09004367
B14=1.10003545
B15=1.10003545
B16=1.09264532
B17=1.09264532
A1=126.28804500
A2=126.28804500
A3=109.12987381
A4=109.12987381
A5=111.03788705
A6=111.03788705
A7=111.52798073
A8=111.52798073
A9=118.68964962
A10=118.68964962
A11=108.26615601
A12=108.26615601
A13=111.48819480
A14=111.48819480
A15=109.57240039
A16=109.57240039
D1=-180.
D2=168.17481779
D3=168.17481779
D4=-88.57827681
D5=-88.57827681
D6=149.17358123
D7=149.17358123
D8=26.03101788
D9=26.03101788
D10=-38.20348554
D11=-38.20348554
D12=81.62214705
D13=81.62214705
D14=-157.48274650
D15=-157.48274650
|
B1=1.22104035
B2=1.38946743
B3=1.38946743
B4=1.45232915
B5=1.45232915
B6=1.09655484
B7=1.09655484
B8=1.0880191
B9=1.0880191
B10=1.44734849
B11=1.44734849
B12=1.08602678
B13=1.08602678
B14=1.09477797
B15=1.09477797
B16=1.08804203
B17=1.08804203
A1=126.26806375
A2=126.26806375
A3=109.29419808
A4=109.29419808
A5=110.89423864
A6=110.89423864
A7=111.55455766
A8=111.55455766
A9=118.74744738
A10=118.74744738
A11=108.20479406
A12=108.20479406
A13=111.38826932
A14=111.38826932
A15=109.82420364
A16=109.82420364
D1=180.
D2=168.43761728
D3=168.43761728
D4=-89.47787724
D5=-89.47787724
D6=148.53156374
D7=148.53156374
D8=26.13643993
D9=26.13643993
D10=-35.32512903
D11=-35.32512903
D12=84.45312914
D13=84.45312914
D14=-154.68170399
D15=-154.68170399
|
|
|
|
|
|
|