|
| |
|
|
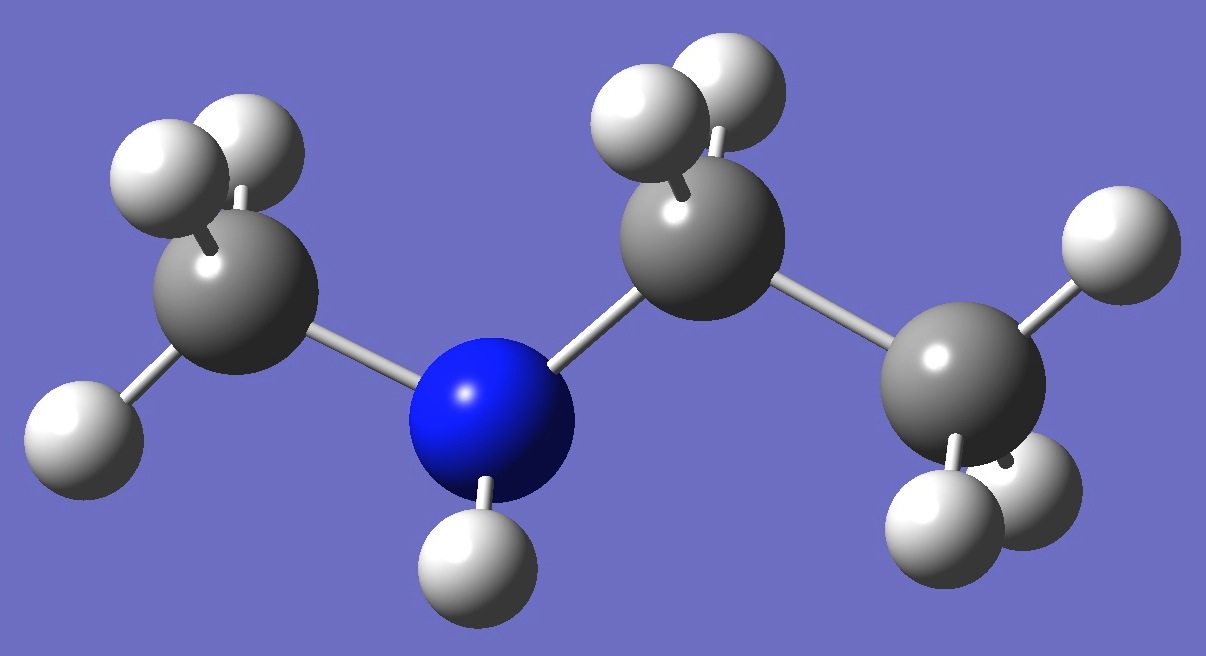
| Table 2. Methylaminoethane. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
mPW1PW91/6-311+G(3df,3pd) and MP2/6-311+G(df,pd)
|
| |
|
|
|
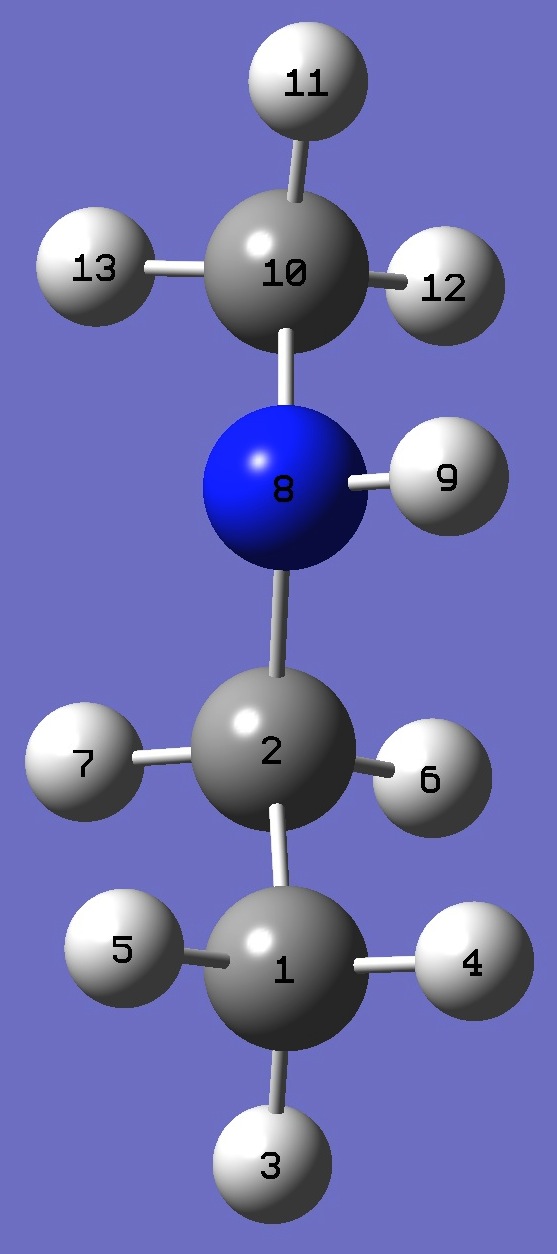
C
C,1,B1
H,1,B2,2,A1
H,1,B3,2,A2,3,D1,0
H,1,B4,2,A3,3,D2,0
H,2,B5,1,A4,3,D3,0
H,2,B6,1,A5,3,D4,0
N,2,B7,1,A6,3,D5,0
H,8,B8,2,A7,1,D6,0
C,8,B9,2,A8,1,D7,0
H,10,B10,8,A9,2,D8,0
H,10,B11,8,A10,2,D9,0
H,10,B12,8,A11,2,D10,0
|
|
|
mPW1PW91
|
MP2 |
|
|
B1=1.51450186
B2=1.08963825
B3=1.09193857
B4=1.08938299
B5=1.10289538
B6=1.09355372
B7=1.44855941
B8=1.01076874
B9=1.44593343
B10=1.08924432
B11=1.10068426
B12=1.09126386
A1=110.88913303
A2=111.04384317
A3=110.6660078
A4=109.65473287
A5=109.68746023
A6=111.29175838
A7=109.47999394
A8=113.40148349
A9=109.90708534
A10=113.93543347
A11=109.51708296
D1=119.52398587
D2=-120.58127068
D3=-56.60369382
D4=59.63793782
D5=178.70837056
D6=54.08043423
D7=177.28257608
D8=-176.50998474
D9=-54.80807661
D10=65.59049757
|
B1=1.5179435
B2=1.09124414
B3=1.09281349
B4=1.09040066
B5=1.10386586
B6=1.09463489
B7=1.45375464
B8=1.01480248
B9=1.45328974
B10=1.09057735
B11=1.10107669
B12=1.09202778
A1=110.81503664
A2=110.62610507
A3=110.14632526
A4=109.75533233
A5=109.8943412
A6=110.82304862
A7=108.75057609
A8=112.4446784
A9=109.71028175
A10=113.52371688
A11=108.93808908
D1=119.62199035
D2=-120.59623927
D3=-57.74674472
D4=59.40021566
D5=178.00973523
D6=55.51212015
D7=176.94298242
D8=-176.63527088
D9=-54.8761526
D10=65.35359957
|
|
|
|
|