| |
||||||||
| Table
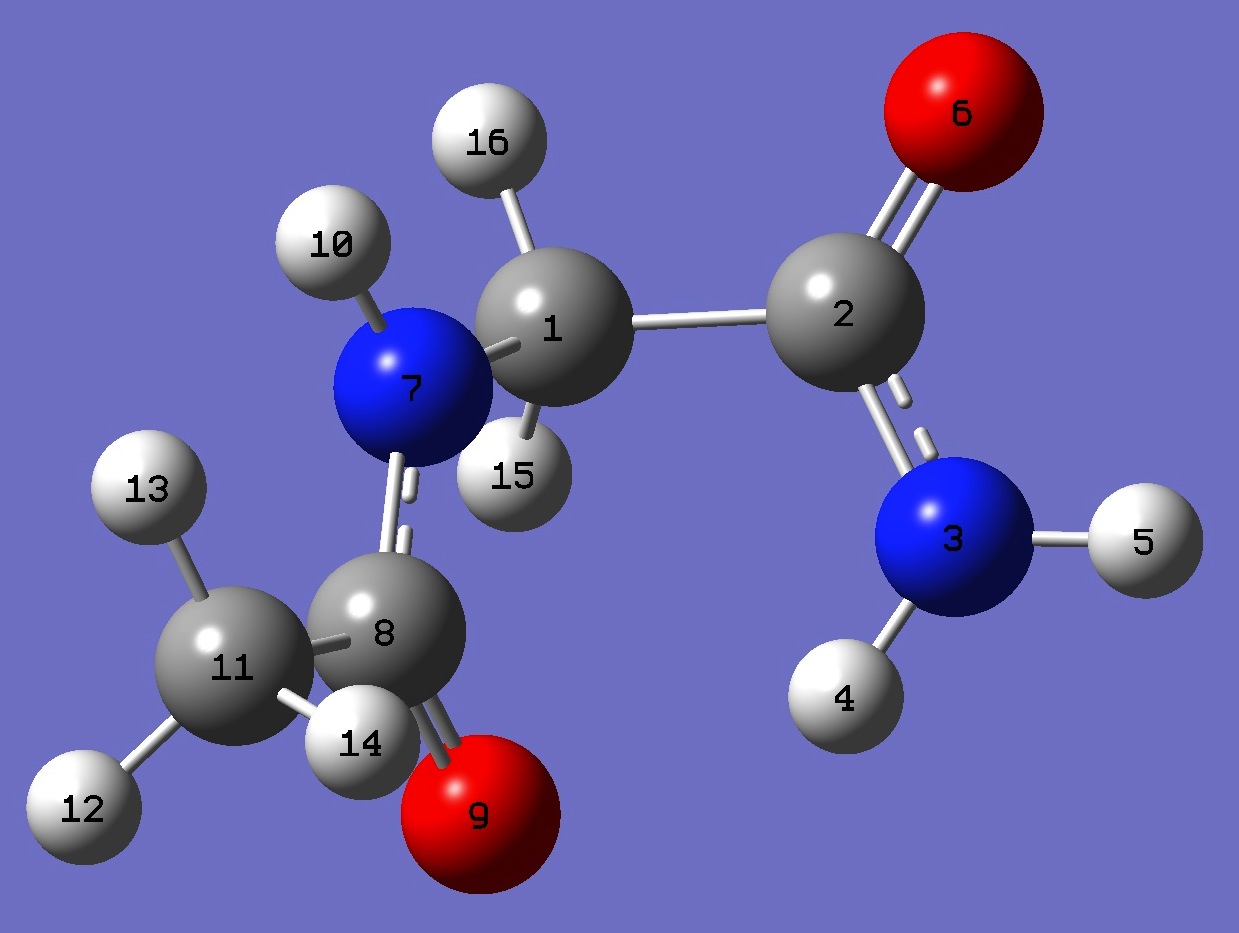
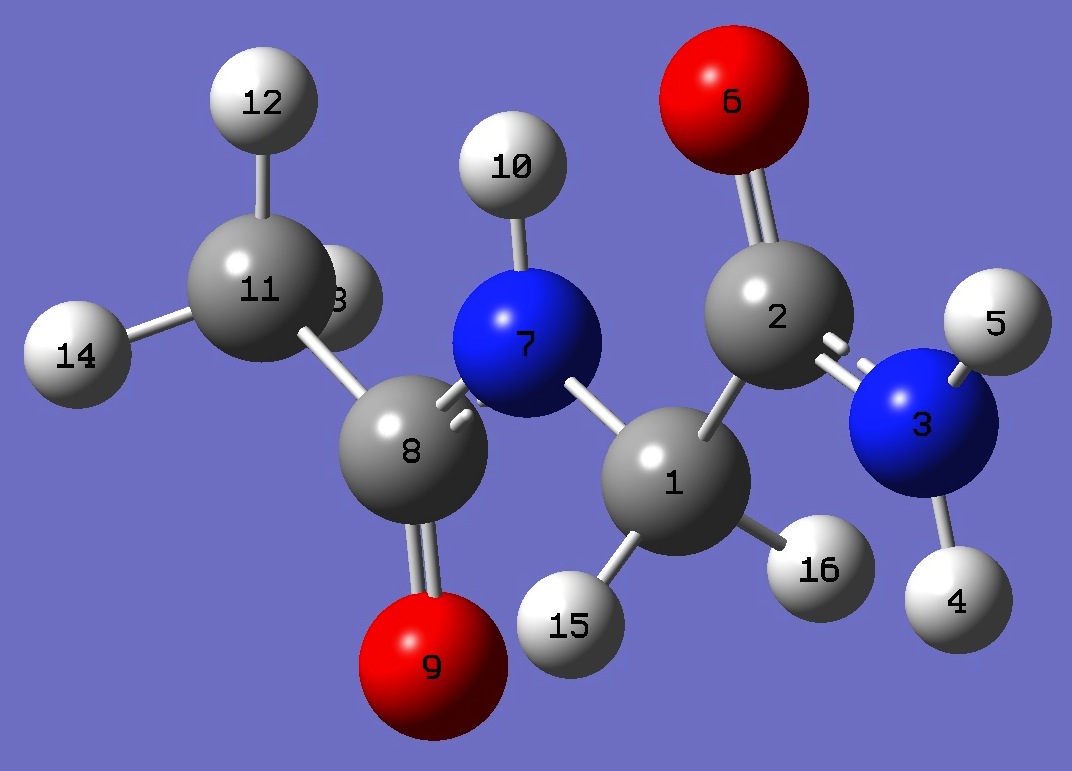
1. 14N nqcc's in C7 N-Acetyl-Glycinamide (MHz). Calculation was made on best estimate re molecular structure. Atomic numbering is shown below in Table 3. |
||||||||
| |
||||||||
| Calc |
Calc [1] |
Expt. [1] |
||||||
| Xaa N(7) | 1.235 |
1.258 |
1.2553(38) |
|||||
| Xbb | 0.970 |
0.752 |
0.717(74) |
|||||
| Xcc | - |
2.206 |
- |
2.008 |
- |
1.972(74) |
||
| Xab | - |
0.744 |
||||||
| Xac | 2.143 |
|||||||
| Xbc | 1.824 |
|||||||
| |
||||||||
| RMS |
0.199 (15. %) |
|||||||
| RSD | 0.030 (1.3 %) |
|||||||
| Xaa N(3) | 0.467 |
0.432 |
0.3887(69) |
|||||
| Xbb | 1.573 | 1.603 | 1.593(10) |
|||||
| Xcc | - |
2.039 |
- |
2.035 |
- |
1.981(10) |
||
| Xab | - |
0.592 |
||||||
| Xac | - |
1.937 |
||||||
| Xbc | - |
1.507 |
||||||
| RMS |
0.057 (4.4 %) |
|||||||
| RSD |
0.030 (1.3 %) | |||||||
| |
||||||||
| |
||||||||
| Table 2. 14N nqcc's in C5 N-Acetyl-Glycinamide (MHz). Calculation was made on best estimate re molecular structure. Atomic numbering is shown below in Table 3. |
||||||||
| |
||||||||
| Calc |
Calc [1] |
Expt. [1] |
||||||
| Xaa N(7) | 2.165 |
2.159 |
2.079(65) |
|||||
| Xbb | 1.887 |
1.748 |
1.787(98) |
|||||
| Xcc | - |
4.053 |
- |
3.907 |
- |
3.865(98) |
||
| Xab | - |
0.532 |
||||||
| |
||||||||
| RMS |
0.133 (5.2 %) |
|||||||
| RSD | 0.030 (1.3 %) |
|||||||
| Xaa N(3) | 2.046 |
1.997 |
2.113(66) |
|||||
| Xbb | 2.064 | 1.923 | 2.08(12) |
|||||
| Xcc | - |
4.110 |
- |
3.920 |
- |
4.19(12) |
||
| Xab | - |
0.077 |
||||||
| RMS |
0.060 (2.2 %) |
|||||||
| RSD |
0.030 (1.3 %) | |||||||
| |
||||||||