|
|
|
|
|
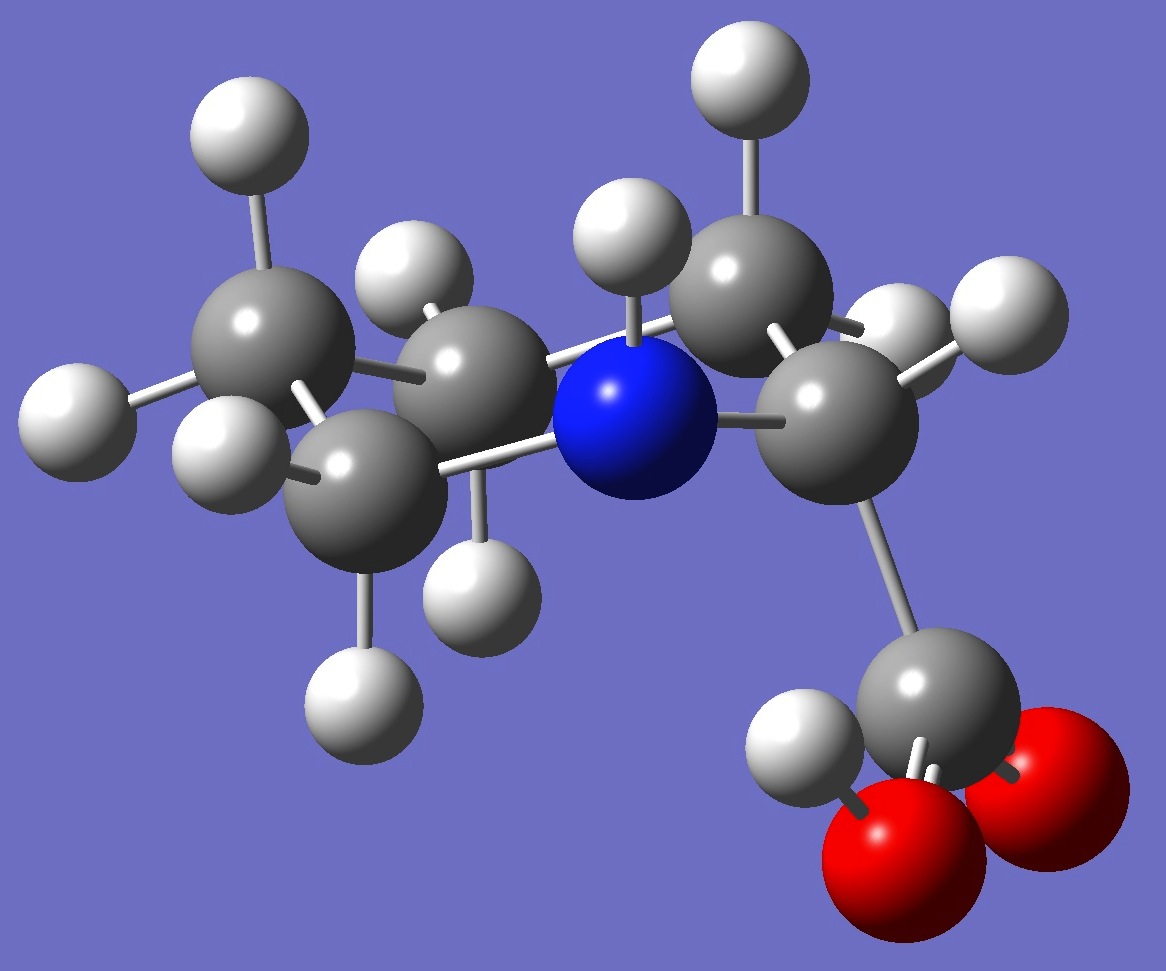
Table 3. Pipecolic Acid B3LYP/cc-pVTZ optimized structure
parameters.
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
N,1,B5,2,A4,3,D3,0
H,6,B6,1,A5,2,D4,0
H,3,B7,2,A6,1,D5,0
H,3,B8,2,A7,1,D6,0
H,1,B9,6,A8,5,D7,0
H,1,B10,6,A9,5,D8,0
H,2,B11,1,A10,6,D9,0
H,2,B12,1,A11,6,D10,0
H,4,B13,3,A12,2,D11,0
H,4,B14,3,A13,2,D12,0
H,5,B15,4,A14,3,D13,0
C,5,B16,4,A15,3,D14,0
O,17,B17,5,A16,4,D15,0
O,17,B18,5,A17,4,D16,0
H,19,B19,17,A18,5,D17,0
|
Conformer I |
|
|
|
|
|
|
|
|
B1=1.52907435
B2=1.53054366
B3=1.53035372
B4=1.53388296
B5=1.4743613
B6=1.01144893
B7=1.09383092
B8=1.09106734
B9=1.09028904
B10=1.09398136
B11=1.09521709
B12=1.09221556
B13=1.09392925
B14=1.088914
B15=1.0941119
B16=1.54245612
B17=1.20070458
B18=1.34231263
B19=0.98163813
A1=110.20682392
A2=110.72346801
A3=112.08627185
A4=114.11986342
A5=110.12855644
A6=109.49507532
A7=110.66957492
A8=107.53887181
|
A9=107.73702102
A10=109.58038351
A11=109.48415969
A12=109.27774531
A13=110.93831662
A14=108.60475588
A15=112.708301
A16=123.92938151
A17=113.2878673
A18=104.99572267
D1=-55.79288218
D2=52.36520198
D3=54.26838484
D4=76.03966423
D5=65.06795447
D6=-177.79553096
D7=-172.64750187
D8=72.6894973
D9=-66.66723037
D10=176.44748682
D11=-67.61621366
D12=174.06353943
D13=-166.18580897
D14=79.33416036
D15=30.34065623
D16=-152.4780923
D17=4.90448485
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
N,1,B5,2,A4,3,D3,0
H,6,B6,1,A5,2,D4,0
H,3,B7,2,A6,1,D5,0
H,3,B8,2,A7,1,D6,0
H,1,B9,6,A8,5,D7,0
H,2,B10,1,A9,6,D8,0
H,2,B11,1,A10,6,D9,0
H,4,B12,3,A11,2,D10,0
H,4,B13,3,A12,2,D11,0
H,5,B14,4,A13,3,D12,0
H,5,B15,4,A14,3,D13,0
C,1,B16,6,A15,5,D14,0
O,17,B17,1,A16,6,D15,0
O,17,B18,1,A17,6,D16,0
H,19,B19,17,A18,1,D17,0
|
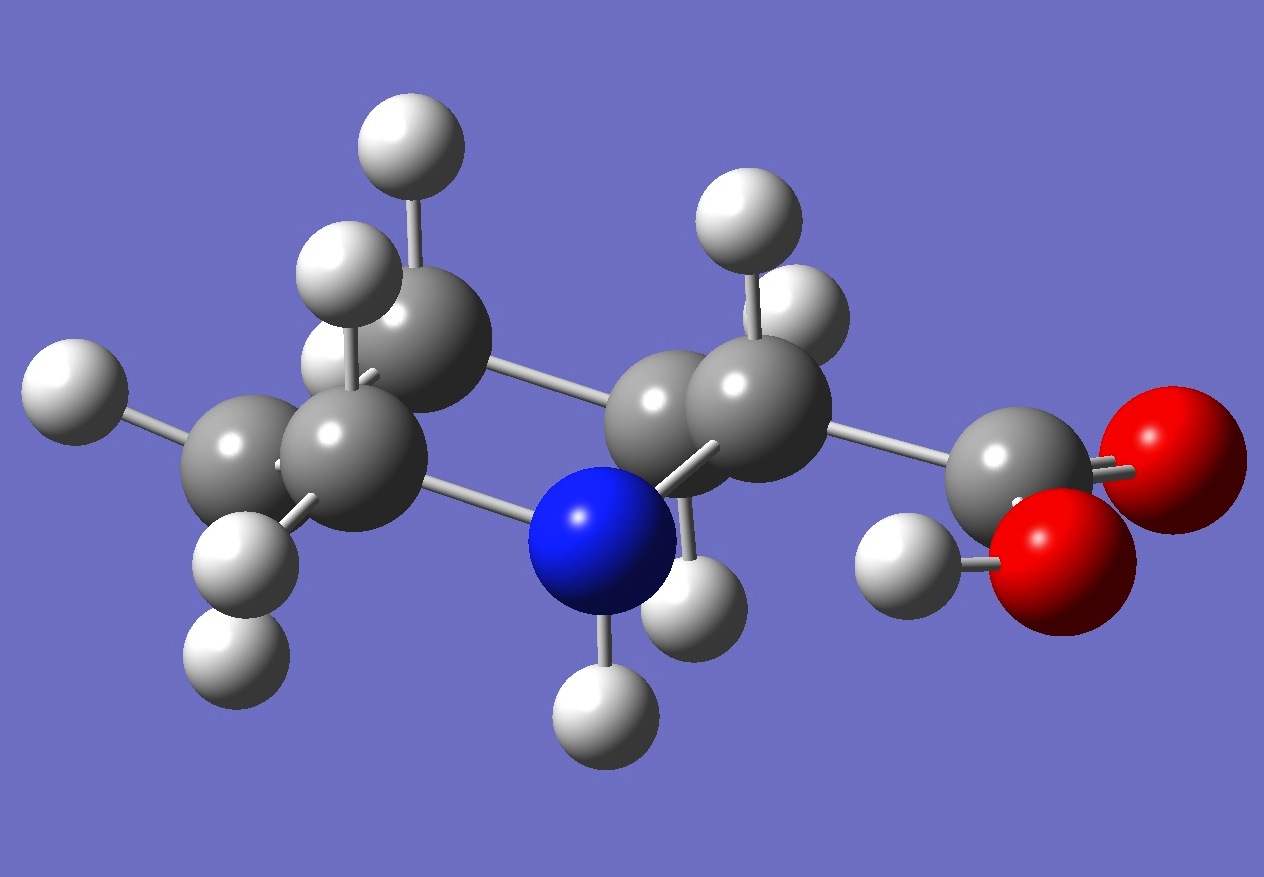
Conformer II
|
|
|
|
|
|
|
|
|
B1=1.52776285
B2=1.53402792
B3=1.53387932
B4=1.53265468
B5=1.47281021
B6=1.01725105
B7=1.09496543
B8=1.09108548
B9=1.09773969
B10=1.0944323
B11=1.08923184
B12=1.09506678
B13=1.09238382
B14=1.0905573
B15=1.09333997
B16=1.53088401
B17=1.19973859
B18=1.34587697
B19=0.97829018
A1=109.67849865
A2=111.7462605
A3=111.45708573
A4=113.41185216
A5=108.29171962
A6=108.96451886
A7=110.19288969
A8=105.92365581
|
A9=108.98584751
A10=109.87536616
A11=109.4829706
A12=110.54779143
A13=111.02318397
A14=110.13419284
A15=109.84858596
A16=124.00935465
A17=113.5163225
A18=106.11968441
D1=-51.99241915
D2=51.30405668
D3=55.43371027
D4=61.37280582
D5=68.71665808
D6=-174.90576205
D7=61.18299748
D8=-65.43390988
D9=178.07347626
D10=-69.11681769
D11=173.75066152
D12=-174.39052844
D13=67.30492034
D14=173.79021747
D15=156.78109878
D16=-25.29285906
D17=3.85905368
|
|
|
|
|
|
|
|