|
| |
|
|
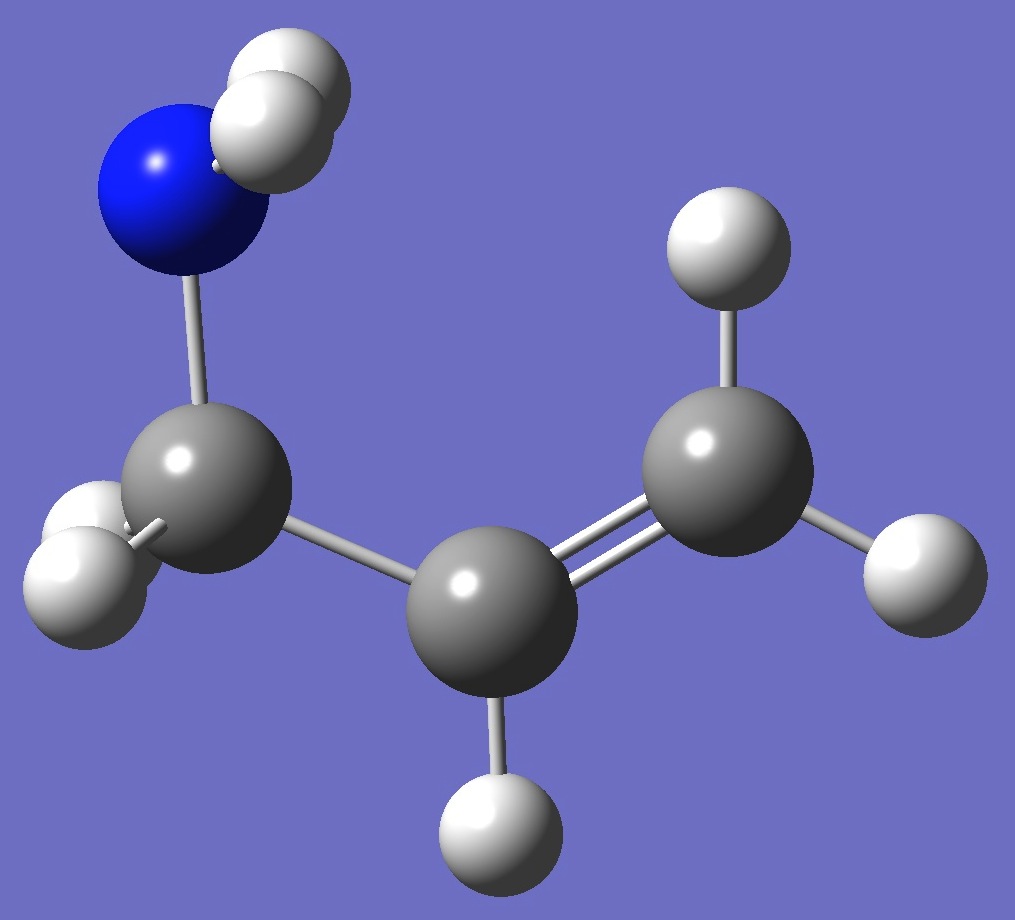
Table 2. Allylamine. Optimized structure parameters
(Å
and degrees). MP2/aug-cc-pVTZ and
MP2/6-311+G(3df,3pd).
|
| |
|
|
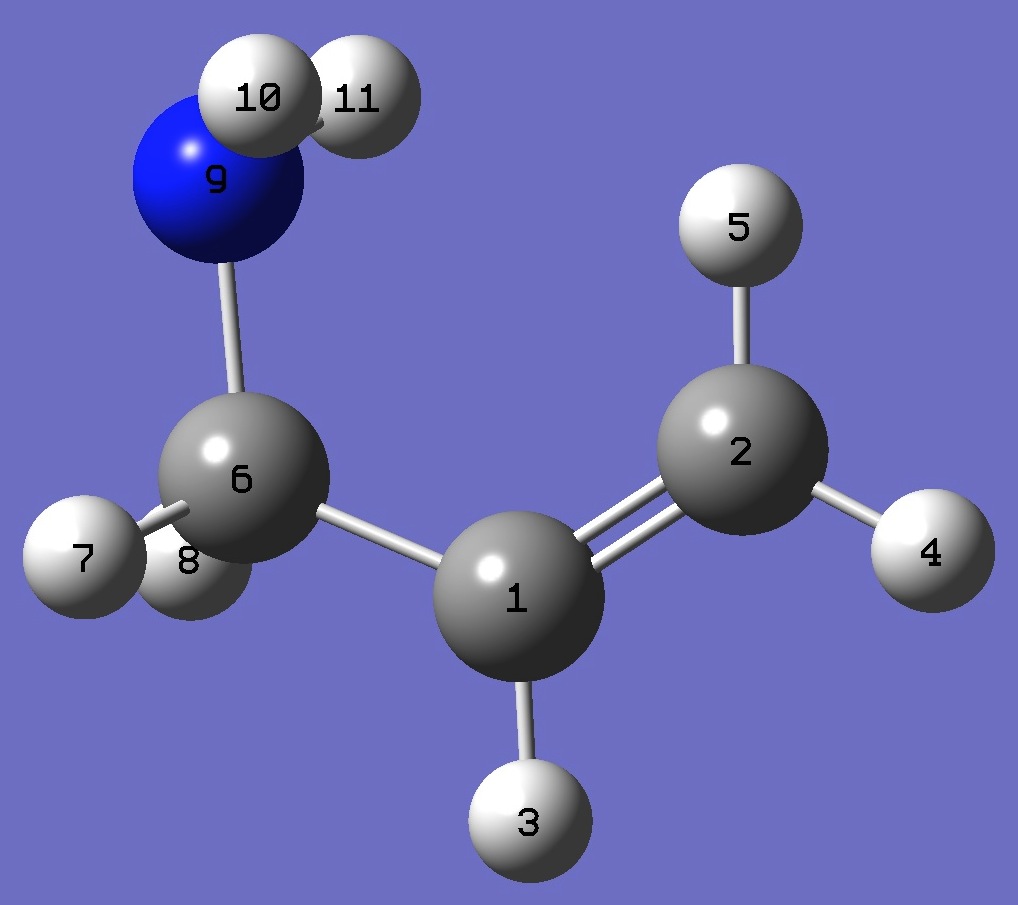
|
C
C,1,B1
H,1,B2,2,A1
H,2,B3,1,A2,3,D1,0
H,2,B4,1,A3,3,D2,0
C,1,B5,2,A4,5,D3,0
H,6,B6,1,A5,2,D4,0
H,6,B7,1,A6,2,D5,0
N,6,B8,1,A7,2,D6,0
H,9,B9,6,A8,1,D7,0
H,9,B10,6,A9,1,D8,0
|
|
|
|
MP2/aug-cc-pVTZ
|
MP2/6-311+G(3df,3pd)
|
|
|
B1=1.33479904
B2=1.08613858
B3=1.08043029
B4=1.08181982
B5=1.50423498
B6=1.09296552
B7=1.09296552
B8=1.45520833
B9=1.01329459
B10=1.01329459
A1=118.95215341
A2=121.00312763
A3=121.53930387
A4=125.45212216
A5=108.56870035
A6=108.56870035
A7=117.5837412
A8=110.23387075
A9=110.23387075
D1=0.
D2=180.
D3=0.
D4=-122.92476799
D5=122.92476799
D6=0.
D7=-58.68940646
D8=58.68940646
|
B1=1.3338801
B2=1.08603383
B3=1.08007344
B4=1.08126857
B5=1.50421458
B6=1.09225238
B7=1.09225238
B8=1.45370872
B9=1.01265992
B10=1.01265992
A1=118.94350171
A2=121.04626124
A3=121.51415459
A4=125.47008543
A5=108.61130549
A6=108.61130549
A7=117.47810239
A8=110.27596937
A9=110.27596937
D1=0.
D2=180.
D3=0.
D4=-122.87982875
D5=122.87982875
D6=0.
D7=-58.73795251
D8=58.73795251
|
|
|