|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
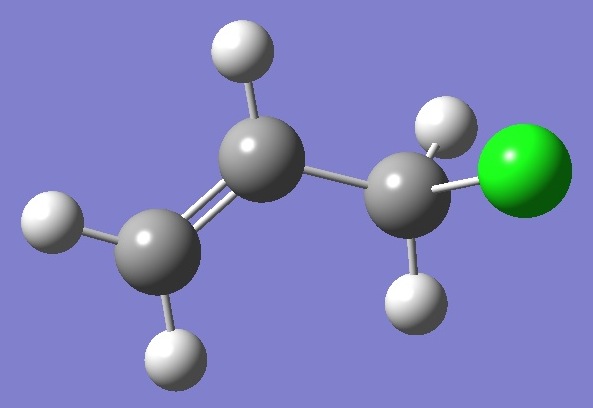
CH2Cl-CH=CH2
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Chlorine |
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
in gauche-3-Chloropropene
|
|
|
|
|
(Allyl
Chloride, skew) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
35Cl nqcc's
in g-3-chloropropene were first determined by Hirota [1]. 35Cl
and 37Cl nqcc's were subsequently determined by Kent et al. [2]. |
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the nqcc tensor
was made here on molecular structures given by MP2/aug-cc-pVTZ(G03) and
MP2/6-311+G(3df,3pd) optimizations; and on these
same structures but with ~re C-C, C=C, and CCl bond
lengths. These calculated nqcc's are compared
with the experimental values [2] in Tables 1 - 4. Structure
parameters are given in Table 5, rotational constants in Table 6. |
|
|
|
|
|
|
|
|
|
|
|
|
In Tables 1 - 4, subscripts a,b,c
refer to the principal axes of the inertia tensor; x,y,z to the
principal axes of the nqcc tensor. Øz,CCl
(degrees) is the angle between the z-principal axis and the CCl bond
direction. ETA = (Xxx - Xyy)/Xzz. |
|
|
RMS is the root mean
square
difference between calculated and experimental diagonal nqcc's
(percentage of the average of the magnitudes of the experimental
nqcc's). RSD is the calibration residual standard deviation of
the B1LYP/TZV(3df,2p) model for calculation of the nqcc's, which may be
taken as an estimate of the uncertainty in the calculated nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 1. 35Cl
nqcc's in g-3-Chloropropene (MHz). |
|
|
|
|
|
|
|
|
|
|
|
Calc (1) was made on the
MP2/aug-cc-pVTZ(G03) optimized molecular structure. |
|
|
Calc (2) was made on this same
structure but with ~re C-C, C=C, and CCl bond lengths. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc (1) |
|
Calc (2) |
|
Expt. [2] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
39.23 |
- |
39.13 |
- |
39.5(5) |
|
|
Xbb |
|
3.00 |
|
2.95 |
|
3.5(4) |
|
|
Xcc |
|
36.23 |
|
36.18 |
|
36.1(6) |
|
|
|Xab| |
|
50.44 * |
|
50.38 * |
|
|
|
|
|Xac| |
|
2.67 |
|
2.75 |
|
|
|
|
|Xbc| |
|
0.55 |
|
0.60 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.34 (1.3 %)
|
|
0.38 (1.5 %)
|
|
|
|
|
RSD |
|
0.49 (1.1 %) |
|
0.49 (1.1 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
37.46 |
|
37.40 |
|
|
|
|
Xyy |
|
35.40 |
|
35.35 |
|
|
|
|
Xzz |
- |
72.86 |
- |
72.75 |
|
|
|
|
ETA |
- |
0.0283 |
- |
0.0283 |
|
|
|
|
Øz,CCl |
|
0.34 |
|
0.35 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* The algabraic sign of the product XabXacXbc is
negative. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 2. 37Cl
nqcc's in g-3-Chloropropene (MHz). |
|
|
|
|
|
|
|
|
|
|
|
Calc (1) was made on the
MP2/aug-cc-pVTZ(G03) optimized molecular structure. |
|
|
Calc (2) was made on this same
structure but with ~re C-C, C=C, and CCl bond lengths. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc (1) |
|
Calc (2) |
|
Expt. [2] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
31.21 |
- |
31.13 |
- |
31.6(8) |
|
|
Xbb |
|
2.65 |
|
2.61 |
|
3.1(10) |
|
|
Xcc |
|
28.56 |
|
28.52 |
|
28.5(13) |
|
|
|Xab| |
|
39.64 * |
|
39.59 * |
|
|
|
|
|Xac| |
|
1.91 |
|
1.98 |
|
|
|
|
|Xbc| |
|
0.30 |
|
0.34 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.34 (1.6 %) |
|
0.39 (1.9 %) |
|
|
|
|
RSD |
|
0.44 (1.1 %) |
|
0.44 (1.1 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* The algabraic sign of the product XabXacXbc is
negative. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 3. 35Cl
nqcc's in g-3-Chloropropene (MHz). |
|
|
|
|
|
|
|
|
|
|
|
Calc (1) was made on the
MP2/6-311+G(3df,3pd) optimized molecular structure. |
|
|
Calc (2) was made on this same
structure but with ~re C-C, C=C, and CCl bond lengths. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc (1) |
|
Calc (2) |
|
Expt. [2] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
38.76 |
- |
38.94 |
- |
39.5(5) |
|
|
Xbb |
|
2.61 |
|
2.72 |
|
3.5(4) |
|
|
Xcc |
|
36.15 |
|
36.22 |
|
36.1(6) |
|
|
|Xab| |
|
50.54 * |
|
50.58 * |
|
|
|
|
|Xac| |
|
1.81 |
|
1.80 |
|
|
|
|
|Xbc| |
|
0.02 |
|
0.01 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.67 (2.5 %) |
|
0.56 (2.1 %) |
|
|
|
|
RSD |
|
0.49 (1.1 %) |
|
0.49 (1.1 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
37.39 |
|
37.44 |
|
|
|
|
Xyy |
|
35.31 |
|
35.38 |
|
|
|
|
Xzz |
- |
72.70 |
- |
72.83 |
|
|
|
|
ETA |
- |
0.0286 |
- |
0.0282 |
|
|
|
|
Øz,CCl |
|
0.36 |
|
0.36 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
* The algabraic sign of the product XabXacXbc is
positive. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 4. 37Cl
nqcc's in g-3-Chloropropene (MHz). |
|
|
|
|
|
|
|
|
|
|
|
Calc (1) was made on the
MP2/6-311+G(3df,3pd) optimized molecular structure. |
|
|
Calc (2) was made on this same
structure but with ~re C-C, C=C, and CCl bond lengths. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc (1) |
|
Calc (2) |
|
Expt [2]
|
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
30.84 |
- |
30.98 |
-
|
31.6(8)
|
|
|
Xbb |
|
2.35 |
|
2.44 |
|
3.1(10)
|
|
|
Xcc |
|
28.49 |
|
28.54 |
|
28.5(13)
|
|
|
|Xab| |
|
39.71 * |
|
39.74 * |
|
|
|
|
|Xac| |
|
1.25 |
|
1.24 |
|
|
|
|
|Xbc| |
|
0.14 |
|
0.13 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS
|
|
0.61 (2.9 %)
|
|
0.52 (2.5 %)
|
|
|
|
|
RSD |
|
0.44 (1.1 %) |
|
0.44 (1.1 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* The algabraic sign of the product XabXacXbc is
positive. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Table 5.
g-3-Chloropropene. Heavy atom structure parameters (Å and degrees).
Complete structures are given here
in Z.matrix format. |
| |
|
|
|
|
r (1) = MP2/aug-cc-pVTZ(G03)
opt. |
|
r (2) = r (1) but
with ~re C-C, C=C, and CCl
bond lengths. |
| |
|
|
|
| Point Group C1 |
|
r (1) |
r (2) |
|
|
|
|
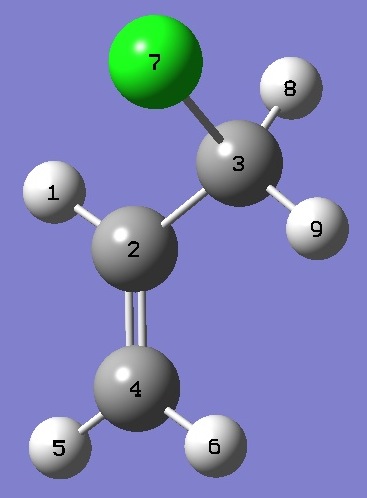
|
ClC(3) |
1.7988 |
1.7956 |
| C(3)C(2) |
1.4862 |
1.4857 |
| C(2)C(4) |
1.3349 |
1.3311 |
| ClC(3)C(2) |
110.41 |
110.41 |
| C(3)C(2)C(4) |
122.83 |
122.83 |
| ClC(3)C(2)C(4) |
118.23 |
118.23 |
| |
|
|
| r (1) = MP2/6-311+G(3df,3pd)
opt. |
| r (2) = r (1) but with MP2/6-311+G(3df,3pd) ~re C-C,
C=C, and CCl bond lengths. |
|
|
|
|
r (1) |
r (2) |
|
|
|
| ClC(3) |
1.7891 |
1.7937 |
| C(3)C(2) |
1.4871 |
1.4866 |
| Click on image to enlarge. |
C(2)C(4) |
1.3338 |
1.3308 |
|
ClC(3)C(2) |
110.59 |
110.59 |
|
C(3)C(2)C(4) |
122.94 |
122.94 |
|
ClC(3)C(2)C(4) |
117.32 |
117.32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Table 6.
g-3-Chloropropene. Rotational constants (MHz). 35Cl
species. |
| |
|
|
|
|
|
r (1) = MP2/aug-cc-pVTZ(G03)
opt. |
|
r (2) = r (1) but
with ~re C-C, C=C, and CCl bond lengths. |
|
|
|
|
|
|
|
r (1) |
r (2) |
Expt. [1] |
|
|
|
|
|
|
A |
21 315.8 |
21 365.8 |
21 669.09(33) |
|
B |
2 831.0 |
2 840.0 |
2 800.902(56) |
|
C |
2 743.7 |
2 751.9 |
2 713.990(55) |
| |
|
|
|
|
|
r (1) = MP2/6-311+G(3df,3pd)
opt. |
|
r (2) = r (1) but with ~re
C-C, C=C, and CCl bond length. |
|
|
|
|
|
|
|
r (1) |
r (2) |
Expt. [1] |
|
|
|
|
|
|
A |
21 263.6 |
21 251.1 |
21 669.09(33) |
|
B |
2 845.8 |
2 843.0 |
2 800.902(56) |
|
C |
2 758.4 |
2 755.1 |
2 713.990(55) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] E.Hirota, J.Mol.Spectrosc.
35,9(1970): Xaa = 39.42, Xbb = 3.45, and Xcc = 35.98 MHz
|
|
|
[2] E.B.Kent, M.N.McCabe,
M.A.Phillips, B.P.Gorden, and S.T.Shipman, 66th OSU International
Symposium on Molecular Spectroscopy, 2011, Abstract RH01. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
cis-3-Chloropropene |
gauche-3-Bromopropene
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Chlorine |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
gCH2ClCHCH2.html |
|
|
|
|
|
|
Last
Modified 7 April 2008 |
|
|
|
|
|
|
|
|
|
|

