|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
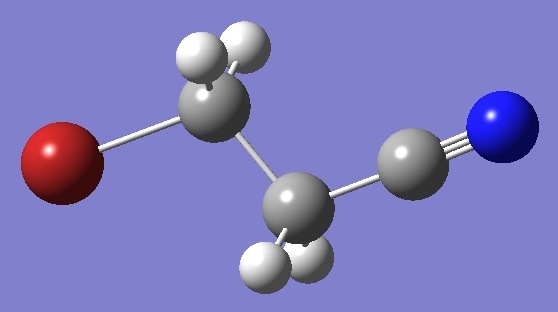
CH2BrCH2CN |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Bromine and Nitrogen
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
in trans 3-Bromopropionitrile
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the bromine and nitrogen nqcc tensors in the trans (Cs) conformer of 3-bromopropionitrile was made on a structure derived ab initio
as discussed below. These are compared with the experimental
bromine nqcc's [1] in Tables 1 and 2. Calculated nitrogen nqcc's
are given in Tables 3 and 4. No experimental nqcc's for nitrogen
have been reported. Structure parameters are given in
Table 5, rotational constants in
Table 6. |
|
|
|
|
|
|
|
|
|
|
|
|
In Tables 1 - 4, subscripts a,b,c refer to the principal axes of the inertia
tensor, subscripts x,y,z to the principal axes of the nqcc tensor. Ø (degrees)
is the angle between its subscripted parameters. ETA = (Xxx
- Xyy)/Xzz. |
|
|
RMS is the root mean square
difference between calculated and experimental nqcc's (percentage of
the average experimental nqcc). RSD is the residual standard deviation
of calibration of the model for calculation of
the nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 1. 79Br nqcc's in trans CH2BrCH2CN (MHz). |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
79Br |
Xaa |
|
487.11 |
|
413(6) |
|
|
|
Xbb |
- |
197.64 |
- |
194 * |
|
|
|
Xcc |
- |
289.47 |
- |
219 * |
|
|
|
|Xab| |
|
283.86 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
59 (21 %) |
|
|
|
|
|
RSD |
|
1.58 (0.39 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
- |
281.49 |
|
|
|
|
|
Xyy |
- |
289.47 |
|
|
|
|
|
Xzz |
|
570.96 |
|
|
|
|
|
ETA |
|
0.0140 |
|
|
|
|
|
Øz,a |
|
18.28 |
|
|
|
|
|
Øa,CBr |
|
17.90 |
|
|
|
|
|
Øz,CBr |
|
0.38 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* Calculated here from the experimental Xaa and (Xbb - Xcc)/Xaa = 0.061(110) [1]. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 2. 81Br nqcc's in trans CH2BrCH2CN (MHz). |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
81Br |
Xaa |
|
407.00 |
|
349(6) |
|
|
|
Xbb |
- |
165.16 |
- |
183 * |
|
|
|
Xcc |
- |
241.84 |
- |
166 * |
|
|
|
|Xab| |
|
212.05 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
56 (24 %) |
|
|
|
|
|
RSD |
|
1.38 (0.40 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
- |
238.18 |
|
|
|
|
|
Xyy |
- |
241.84 |
|
|
|
|
|
Xzz |
|
477.02 |
|
|
|
|
|
ETA |
|
0.0140 |
|
|
|
|
|
Øz,a |
|
18.27 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* Calculated here from the experimental Xaa and (Xbb - Xcc)/Xaa = - 0.048(67) [1]. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 3. 14N nqcc's
in trans CH279BrCH2CN (MHz). |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
- |
3.577 |
|
|
|
|
|
Xbb |
|
1.554 |
|
|
|
|
|
Xcc |
|
2.023 |
|
|
|
|
|
|Xab| |
|
2.059 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
2.278 |
|
|
|
|
|
Xyy |
|
2.023 |
|
|
|
|
|
Xzz |
- |
4.301 |
|
|
|
|
|
ETA |
- |
0.059 |
|
|
|
|
|
Øz,a |
|
19.37 |
|
|
|
|
|
Øa,CN |
|
19.75 |
|
|
|
|
|
Øz,CN |
|
0.38 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 4. 14N nqcc's
in trans CH281BrCH2CN (MHz). |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
- |
3.577 |
|
|
|
|
|
Xbb |
|
1.554 |
|
|
|
|
|
Xcc |
|
2.023 |
|
|
|
|
|
|Xab| |
|
2.059 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Molecular Structure |
|
|
|
|
|
|
|
|
|
|
|
|
The molecular structure was optimized
at the MP2/6-311+G(d,p) level of theory. The optimized C-C,
C-C(N), and CN bond lengths were corrected using equations obtained
from linear regression analyses of the data given in Tables VIII and IX
of Ref. [2]. The CH bond
lengths were corrected using r = 1.001 ropt, where ropt
is obtained by MP2/6-31G(d,p) optimization [3]. Interatomic
angles used in the calculation are those given by MP2/6-311+G(2d,p)
optimization. |
|
|
For the C-Br bond length,
optimization was made at the MP2/6-311+G(2d,p) level of theory of the
C-Br equilibrium bond lengths in CH3Br, CH2Br2,
HCCBr, and BrCN. Linear regression of the calculated versus
equilibrium bond lengths yields the following relationship, by which
the C-Br was corrected: |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
r = 0.9946 × ropt + 0.0001, RSD = 0.0015 Å. |
|
|
|
|
|
|
|
|
|
|
|
|
The optimized C-Br bond length is 1.9442 Å which, after correction is 1.934 Å. |
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
| Table 5.
trans 3-Bromopropionitrile. Heavy atom structure parameters
(Å and degrees). The complete structure is given here in Z-Matrix format. |
| |
|
|
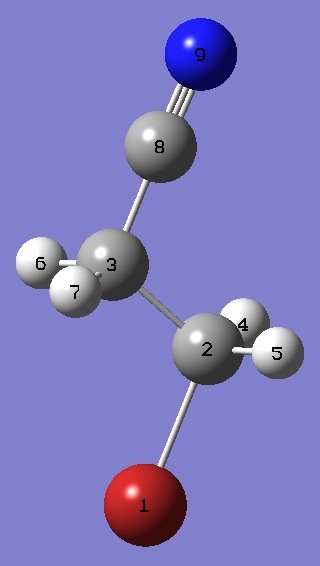
|
NC(8) |
1.156 |
| C(8)C(3) |
1.462 |
| C(3)C(2) |
1.521 |
| C(2)Br |
1.934 |
| NC(8)C(3) |
178.43 |
| C(8)C(3)C(2) |
109.98 |
| C(3)C(2)Br |
110.26 |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
| Table 6. trans 3-Bromopropionitrile. Rotational constants (MHz). Normal species. |
| |
|
|
|
|
|
Calc. ropt |
Expt. [1] |
|
|
|
|
|
A |
26 950.2 |
26 251.22(77.73) |
|
B |
1 037.8 |
1 033.69(1) |
|
C |
1 011.9 |
1 007.76(1) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] S.Xu and M.D.Harmony, J.Mol.Struct. 274,115(1992). |
|
|
[2] J.Demaison, J.Cosléou, R.Bocquet, and A.G.Lesarri, J.Mol.Spectrosc. 167,400(1994). |
|
|
[3] J.Demaison and G.Wlodarczak, Structural Chem. 5,57(1994). |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
t-3-Chloropropionitrile |
gau-3-Chloropropionitrile |
|
|
|
|
Methyl Bromide |
Ethyl Bromide |
|
|
|
|
2-Bromopropane |
2-Bromoethanol |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Bromine |
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
tCH2BrCH2CN.html |
|
|
|
|
|
|
Last
Modified 9 July 2006 |
|
|
|
|
|
|
|
|
|
|

