|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
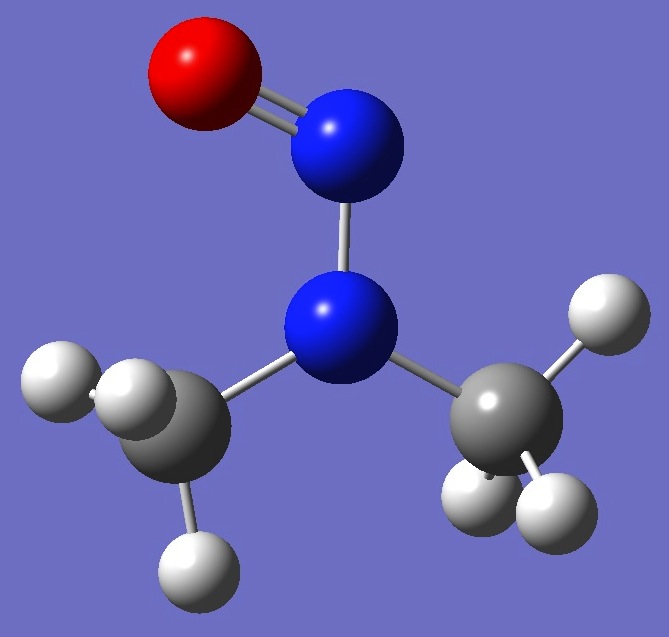
(CH3)2NN=O |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
|
in Dimethylnitrosamine |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the
nitrogen nqcc's
in dimethylnitrosamine was made here on a molecular structure given by
B3LYP/cc-pVTZ optimization. These are compared with the
experimental nqcc's of Heineking and Dreizler [1] in Tables 1 and 2;
and those of Guarnieri et al. [2] in Tables 3 and 4. Structure
parameters are
given in Z-matrix format in Table 5, rotational
constants in Table 6. |
|
|
|
|
|
|
|
|
|
|
|
|
In Tables 1 - 4, subscripts a,b,c
refer to the principal axes of the inertia tensor; x,y,z to the
principal axes of the nqcc tensor. ETA = (Xxx - Xyy)/Xzz. Ø (degrees) is the angle between its
subscripted parameters.
|
|
|
RMS is the root mean square differene between calculated and
experimental nqcc's (percent of average experimental nqcc). RSD
is the calibration residual standard deviation of the B3PW91/6-311+G(df,pd) model for
calculation of the efg's/nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 1. Amino nitrogen nqcc's in (CH3)2NN=O (MHz). Calculation
was made on the B3LYP/cc-pVTZ ropt molecular structure. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.986
|
|
2.0210(25)
|
|
|
|
Xbb |
|
2.107
|
|
2.1287(31)
|
|
|
|
Xcc |
-
|
4.092
|
-
|
4.1497(31)
|
|
|
|
|Xab| |
|
0.470
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.033 (1.2 %)
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
1.572
|
|
|
|
|
|
Xyy |
|
2.520
|
|
|
|
|
|
Xzz |
-
|
4.092
|
|
|
|
|
|
ETA |
|
0.232
|
|
|
|
|
|
Øz,c |
|
0
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 1. Nitroso nitrogen nqcc's in (CH3)2NN=O (MHz). Calculation
was made on the B3LYP/cc-pVTZ ropt molecular structure. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.870
|
|
1.9054(25)
|
|
|
|
Xbb |
-
|
5.482
|
-
|
5.4118(27)
|
|
|
|
Xcc |
|
3.612
|
|
3.5064(27)
|
|
|
|
|Xab| |
|
1.346
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.075 (2.1 %)
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
2.108
|
|
|
|
|
|
Xyy |
|
3.612
|
|
|
|
|
|
Xzz |
-
|
5.720
|
|
|
|
|
|
ETA |
|
0.263
|
|
|
|
|
|
Øz,a |
|
79.94
|
|
|
|
|
|
Øa,bi* |
|
87.26
|
|
|
|
|
|
Øz,bi |
|
7.32
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* "bi" is bisector of NN=O angle.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 3. Amino nitrogen nqcc's in (CH3)2N15N=O (MHz). Calculation
was made on the B3LYP/cc-pVTZ ropt molecular structure. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [2] |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.994
|
|
1.92(10)
|
|
|
|
Xbb |
|
2.098
|
|
2.09(10)
|
|
|
|
Xcc |
-
|
4.092
|
-
|
4.11(10)
|
|
|
|
|Xab| |
|
0.471
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.04 (1.6 %)
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 4. Nitroso nitrogen nqcc's in (CH3)215NN=O (MHz). Calculation
was made on the B3LYP/cc-pVTZ ropt molecular structure. |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc. |
|
Expt. [2] |
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.870
|
|
1.83(10)
|
|
|
|
Xbb |
-
|
5.482
|
-
|
5.55(10)
|
|
|
|
Xcc |
|
3.612
|
|
3.72(10)
|
|
|
|
|Xab| |
|
1.345
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.08 (2.1 %)
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
| Table 5. Dimethylnitrosamine. B3LYP/cc-pVTZ optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
|
|
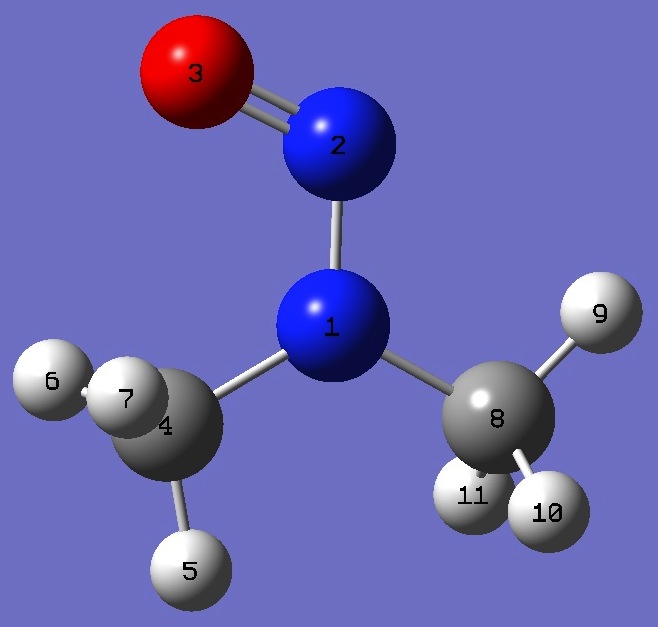
|
N
N,1,B1
O,2,B2,1,A1
C,1,B3,2,A2,3,D1,0
H,4,B4,1,A3,2,D2,0
H,4,B5,1,A4,2,D3,0
H,4,B6,1,A5,2,D4,0
C,1,B7,2,A6,3,D5,0
H,8,B8,1,A7,2,D6,0
H,8,B9,1,A8,2,D7,0
H,8,B10,1,A9,2,D8,0
|
|
|
|
|
|
|
|
|
B1=1.3265921
B2=1.22186221
B3=1.4541258
B4=1.08630965
B5=1.09039556
B6=1.09039556
B7=1.44750646
B8=1.0868103
B9=1.09200323
B10=1.09200323
A1=114.81412723
A2=120.63540955
A3=108.55615551
A4=110.33673004
A5=110.33673004
A6=116.95618512
A7=107.47315665
A8=111.04216193
A9=111.04216193
D1=0.
D2=180.
D3=59.35126677
D4=-59.35126677
D5=180.
D6=0.
D7=119.47072768
D8=-119.47072768
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 6. Dimethylnitrosamine. Rotational Constants
(MHz), Calc = B3LYP/cc-pVTZ. Centrifugal distortion constants (kHz), Calc = B3LYP/cc-pVTZ.
|
| |
|
|
|
|
|
Calc
|
Expt. [1] |
|
|
|
|
|
A |
9050. |
9053.74(15) |
|
B |
4610.
|
4613.91(11)
|
|
C |
3176.
|
3170.33(11)
|
|
|
|
|
|
Delta_J
|
0.686
|
0.7(71)
|
|
Delta_JK
|
8.18
|
--- |
|
Delta_K
|
0.0615
|
21.7(22)
|
|
delta_J
|
0.187
|
1.086(26)
|
|
delta_K
|
1.65
|
14.8(11)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] N.Heineking and H.Dreizler, Z.Naturforsch. 48a,570(1993).
|
|
|
[2] A.Guarnieri, F.Rohwer, and F. Scappini, Z.Naturforsch. 30a,904(1975).
|
|
|
|
|
|
|
|
|
|
|
|
|
A.Guarnieri and R.Nicolaisen, Z.Naturforsch. 34a,620(1979).
|
|
|
F.Scappini, A.Guarnieri, H.Dreizler, and P.Rademacher, Z.Naturforsch. 27a,1329(1972). |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Methyl ethyl nitrosamine
|
Methyl propyl nitrosamine |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents
|
|
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH32NNO.html |
|
|
|
|
|
|
Last
Modified 31 Oct 2015 |
|
|
|
|
|
|
|
|
|
|

