| |
|||||||
| Table 1. Amino nitrogen nqcc's in OMGA comformer (MHz). Calculation was made on the B3LYP/cc-pVTZ ropt molecular structure. | |||||||
| Calc. | Expt. [1] | ||||||
| |
|||||||
| 14N | Xaa | 1.237 |
0.92(2) |
||||
| Xbb | 2.347 |
2.86(3) |
|||||
| Xcc | - |
3.584 |
- |
3.78 |
|||
| Xab | 0.495 |
||||||
| Xac | 1.371 |
||||||
| Xbc | - |
0.393 |
|||||
| RMS | 0.37 (15. %) |
||||||
| RSD | 0.030 (1.3 %) | ||||||
| Xxx | 1.449 |
||||||
| Xyy | 2.538 |
||||||
| Xzz | - |
3.987 |
|||||
| ETA | 0.273 |
||||||
| |
|||||||
| Table 2. Nitroso nitrogen nqcc's in OMGA conformer (MHz). Calculation was made on the B3LYP/cc-pVTZ ropt molecular structure. | |||||||
| Calc. | Expt. [1] | ||||||
| |
|||||||
| 14N | Xaa | 0.984 |
1.39(1) |
||||
| Xbb | - |
4.152 |
- |
4.90(5) |
|||
| Xcc | 3.168 |
3.51 |
|||||
| Xab | 2.738 |
||||||
| Xac | - |
1.047 |
|||||
| Xbc | 1.570 |
||||||
| RMS | 0.53 (16. %) |
||||||
| RSD | 0.030 (1.3 %) | ||||||
| Xxx | 2.096 |
||||||
| Xyy | 3.630 |
||||||
| Xzz | - |
5.726 |
|||||
| ETA | 0.268 |
||||||
| Øz,bi* |
7.11 |
||||||
| |
|||||||
| Table 3. Amino nitrogen nqcc's in OMGG- comformer (MHz). Calculation was made on the B3LYP/cc-pVTZ ropt molecular structure. | |||||||
| Calc. | Expt. [1] | ||||||
| |
|||||||
| 14N | Xaa | 0.394 |
0.37(2) |
||||
| Xbb | 1.063 |
3.26(14) |
|||||
| Xcc | - |
1.458 |
- |
3.63 |
|||
| Xab | 1.488 |
||||||
| Xac | 1.734 |
||||||
| Xbc | - |
1.458 |
|||||
| RSD | 0.030 (1.3 %) | ||||||
| Xxx | 1.426 |
||||||
| Xyy | 2.539 |
||||||
| Xzz | - |
3.966 |
|||||
| ETA | 0.280 |
||||||
| |
|||||||
| Table 4. Nitroso nitrogen nqcc's in OMGG- conformer (MHz). Calculation was made on the B3LYP/cc-pVTZ ropt molecular structure. | |||||||
| Calc. | Expt. [1] | ||||||
| |
|||||||
| 14N | Xaa | 1.280 |
1.42(1) |
||||
| Xbb | - |
0.686 |
- |
4.48(16) |
|||
| Xcc | - |
0.594 |
3.06 |
||||
| Xab | 2.738 |
||||||
| Xac | - |
1.047 |
|||||
| Xbc | 1.570 |
||||||
| RSD | 0.030 (1.3 %) | ||||||
| Xxx | 2.095 |
||||||
| Xyy | 3.633 |
||||||
| Xzz | - |
5.728 |
|||||
| ETA | 0.268 |
||||||
| Øz,bi* |
7.17 |
||||||
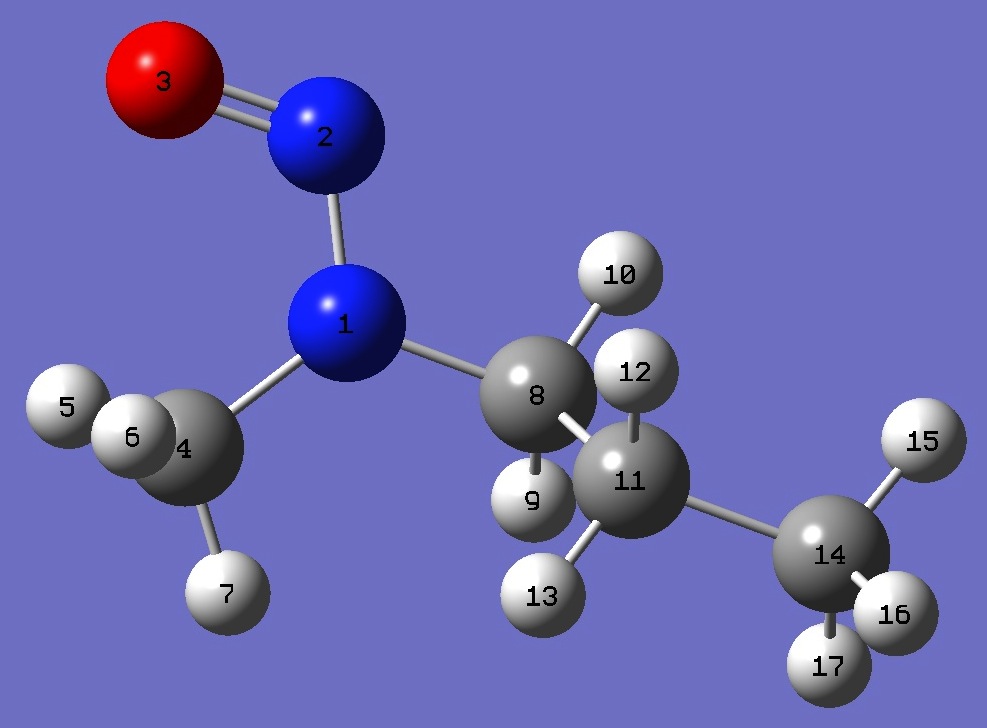
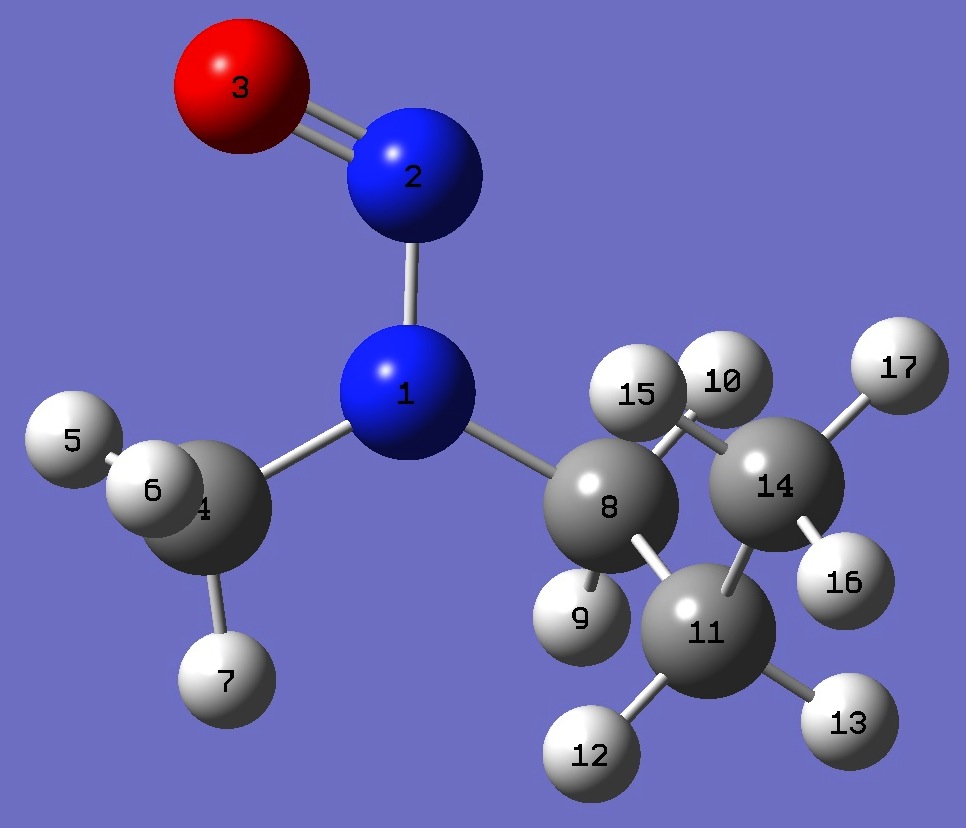
| Table 5. Methylproylnitrosamine. B3LYP/cc-pVTZ optimized molecular structure parameters, ropt (Å and degrees). | ||||
| N N,1,B1 O,2,B2,1,A1 C,1,B3,2,A2,3,D1,0 H,4,B4,1,A3,2,D2,0 H,4,B5,1,A4,2,D3,0 H,4,B6,1,A5,2,D4,0 C,1,B7,2,A6,3,D5,0 H,8,B8,1,A7,2,D6,0 H,8,B9,1,A8,2,D7,0 C,8,B10,1,A9,2,D8,0 H,11,B11,8,A10,1,D9,0 H,11,B12,8,A11,1,D10,0 C,11,B13,8,A12,1,D11,0 H,14,B14,11,A13,8,D12,0 H,14,B15,11,A14,8,D13,0 H,14,B16,11,A15,8,D14,0 |
||||
| OMGA |
OMGG- | |||
| B1=1.32716926 B2=1.22211826 B3=1.45452608 B4=1.0894307 B5=1.09108498 B6=1.08650664 B7=1.4541717 B8=1.09390997 B9=1.08937687 B10=1.53012642 B11=1.0921627 B12=1.09289523 B13=1.52737767 B14=1.09153046 B15=1.08986238 B16=1.09190095 A1=115.02062685 A2=120.4945389 A3=109.61756134 A4=111.08317887 A5=108.63188525 A6=116.67657379 A7=108.74909329 A8=105.91125171 A9=113.80210827 A10=108.53576255 A11=109.66636853 A12=111.91112227 A13=111.23934663 A14=111.07023151 A15=111.37178629 D1=0.47962806 D2=51.77342643 D3=-66.93683843 D4=172.22799663 D5=-177.71352414 D6=-130.09583805 D7=-14.53251905 D8=106.72808485 D9=-58.47591153 D10=57.67936651 D11=179.65854857 D12=59.78638914 D13=179.7173299 D14=-60.31301955 |
B1=1.32791399 B2=1.22213949 B3=1.45432139 B4=1.08942441 B5=1.09117158 B6=1.08642843 B7=1.45531051 B8=1.09313325 B9=1.08947431 B10=1.53214614 B11=1.09329223 B12=1.09304463 B13=1.52692032 B14=1.09033061 B15=1.09025539 B16=1.09124038 A1=115.00075723 A2=120.43459966 A3=109.60935424 A4=111.10921069 A5=108.62916219 A6=116.95662232 A7=108.58153146 A8=105.69920442 A9=114.0082083 A10=109.50213886 A11=107.25514276 A12=113.77090008 A13=111.60777338 A14=110.87743645 A15=111.11622603 D1=1.00198535 D2=50.97753394 D3=-67.7060042 D4=171.47822851 D5=-178.59273892 D6=-126.90970561 D7=-11.33081525 D8=110.11047449 D9=59.87745177 D10=174.4363909 D11=-63.75993105 D12=60.18794415 D13=-179.66241297 D14=-59.70379034 |
|||
| Table 4. Methylpropylnitrosamine. Rotational Constants
(MHz), Calc = B3LYP/cc-pVTZ. |
||||
| Calc |
Expt. [1] | |||
| OMGA |
A | 6349. |
6307.(5) | |
| B | 1297. |
1308.177(6) |
||
| C | 1170. |
1181.171(4) |
||
| OMGG- | A |
4363. | 4254.(6) | |
| B |
1689. | 1748.485(9) | ||
| C |
1490. | 1546.041(13) | ||