| |
|||||||||
| Table
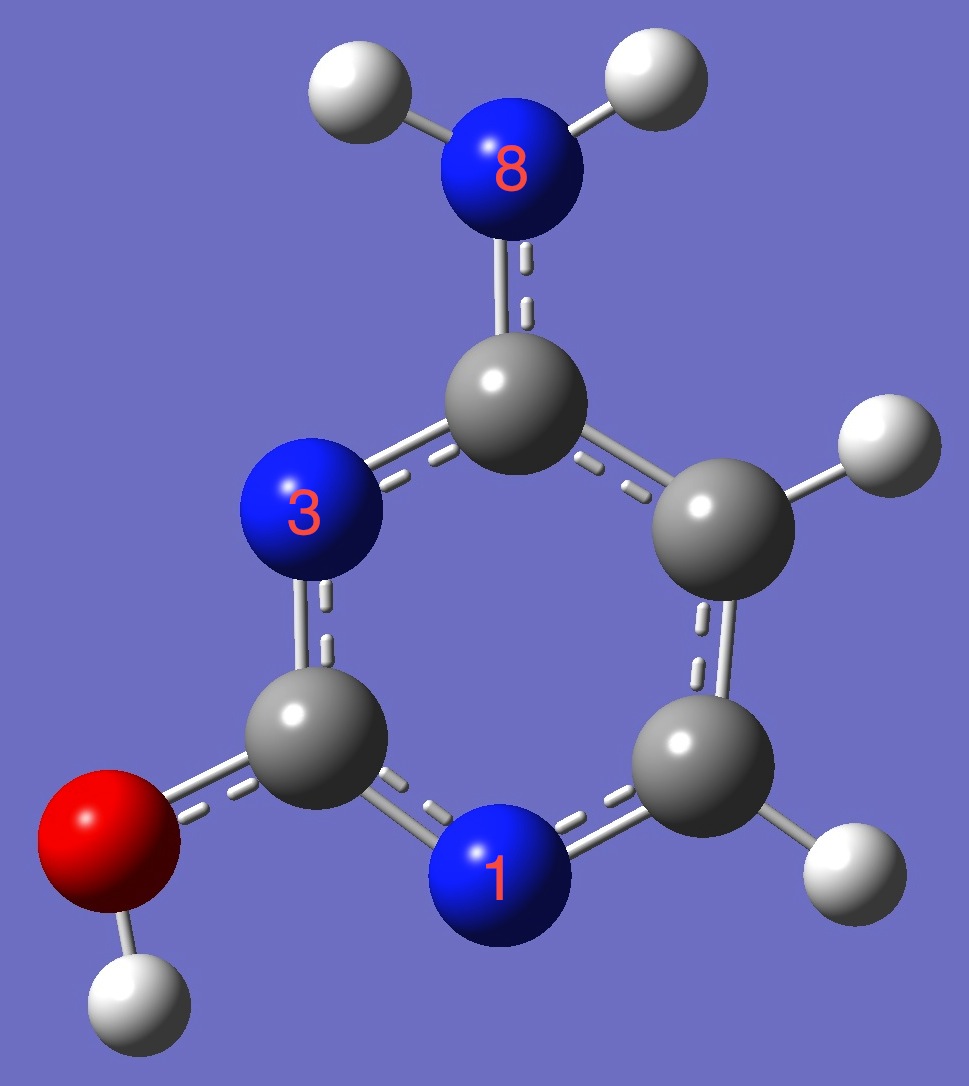
1. 14N nqcc's in trans-Cytosine
(MHz). Calculation was made on molecular structures given by (1)
B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimization. Nitrogen numbering is given above in the figure. |
|||||||||
| |
|||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
|||||||
| N(1) |
Xaa |
- |
2.640 |
- |
2.640 |
- |
2.6373(13) |
||
| Xbb | 1.226 |
1.208 |
1.1672(28) |
||||||
| Xcc | 1.414 |
1.432 |
1.4701(28) |
||||||
| Xab | - |
2.029 |
- |
2.086 |
|||||
| Xac | 0.002 |
0.002 |
|||||||
| Xbc | 0.006 |
0.005 |
|||||||
| |
|||||||||
| RMS |
0.047 (2.7 %) |
0.033(1.8 %) |
|||||||
| N(3) |
Xaa | 2.290 |
2.336 |
2.2619(20) |
|||||
| Xbb | - |
3.624 |
- |
3.672 |
- |
3.6570(22) |
|||
| Xcc | 1.334 |
1.336 |
1.3951(22) |
||||||
| Xab | - |
0.233 |
- |
0.233 |
|||||
| Xac | 0.017 |
0.018 |
|||||||
| Xbc | 0.005 |
0.004 |
|||||||
| RMS | 0.043 (1.8 %) |
0.055 (2.3 %) |
|||||||
| N(8) |
Xaa | 2.087 |
2.199 |
2.2167(17) |
|||||
| Xbb | 1.960 |
2.009 |
1.9511(20) |
||||||
| Xcc | - |
4.047 |
- |
4.208 |
- |
4.1678(20) |
|||
| Xab | 0.160 |
0.179 |
|||||||
| Xac | - |
1.035 |
- |
0.820 |
|||||
| Xbc | - |
0.239 |
- |
0.173 |
|||||
| |
|||||||||
| RMS |
0.102 (3.7 %) |
0.042 (1.5 %) |
|||||||
| |
|||||||||
| |
|||||||||
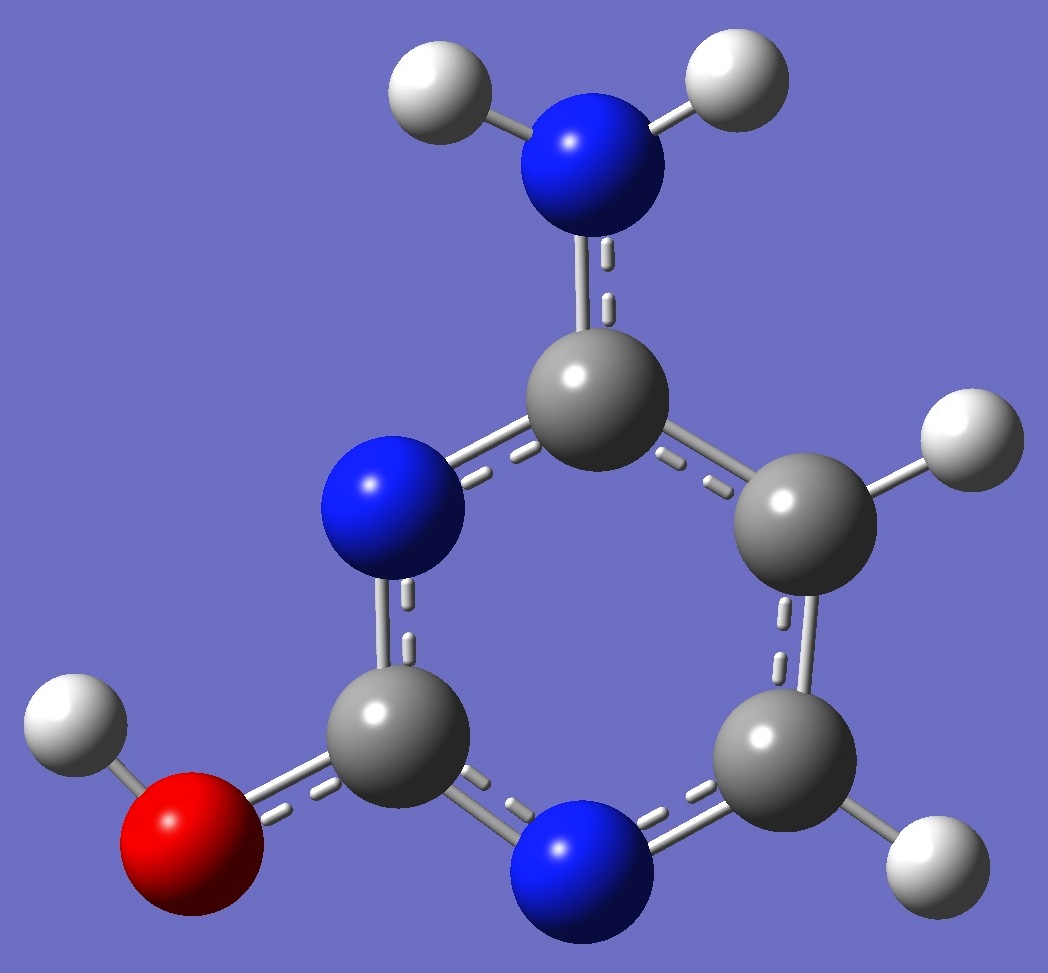
| Table 2. 14N nqcc's in cis-Cytosine (MHz). Calculation was made on molecular structures given by (1) B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimization. Nitrogen numbering is given above in the figure. | |||||||||
| |
|||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
|||||||
| N(1) |
Xaa |
- |
2.822 |
- |
2.818 |
- |
2.8007(18) |
||
| Xbb | 1.074 |
1.063 |
1.0340(27) |
||||||
| Xcc | 1.749 |
1.755 |
1.7667(27) |
||||||
| Xab | - |
2.255 |
- |
2.305 |
|||||
| Xac | 0.002 |
0.002 |
|||||||
| Xbc | 0.003 |
0.003 |
|||||||
| |
|||||||||
| RMS |
0.028 (1.5 %) |
0.021(1.1 %) |
|||||||
| N(3) |
Xaa | 2.273 |
2.319 |
2.2371(23) |
|||||
| Xbb | - |
3.331 |
- |
3.379 |
- |
3.3890(25) |
|||
| Xcc | 1.058 |
1.060 |
1.1519(25) |
||||||
| Xab | - |
0.090 |
- |
0.098 |
|||||
| Xac | 0.014 |
0.016 |
|||||||
| Xbc | 0.002 |
0.001 |
|||||||
| RMS | 0.067 (2.9 %) |
0.071 (3.1 %) |
|||||||
| N(8) |
Xaa | 2.078 |
2.206 |
2.2237(17) |
|||||
| Xbb | 1.989 |
2.051 |
1.9832(20) |
||||||
| Xcc | - |
4.067 |
- |
4.257 |
- |
4.2069(20) |
|||
| Xab | 0.163 |
0.186 |
|||||||
| Xac | - |
1.058 |
- |
0.821 |
|||||
| Xbc | - |
0.314 |
- |
0.225 |
|||||
| |
|||||||||
| RMS |
0.116 (4.2 %) |
0.049 (1.7 %) |
|||||||
| |
|||||||||
| Table 3. Cytosine. B3P86/6-31G(3d,3p) and B3LYP/cc-pVTZ optimized structure parameters (Å and degrees). | |||||
| trans |
|||||
| C N,1,B1 C,2,B2,1,A1 C,3,B3,2,A2,1,D1,0 C,4,B4,3,A3,2,D2,0 N,1,B5,2,A4,3,D3,0 H,3,B6,2,A5,1,D4,0 H,4,B7,3,A6,2,D5,0 O,1,B8,2,A7,3,D6,0 H,9,B9,1,A8,2,D7,0 N,5,B10,4,A9,3,D8,0 H,11,B11,5,A10,4,D9,0 H,11,B12,5,A11,4,D10,0 |
|||||
| B3P86 | B3LYP |
||||
| B1=1.33176409 B2=1.33726757 B3=1.3769012 B4=1.40602979 B5=1.32418979 B6=1.08747424 B7=1.0827569 B8=1.33865157 B9=0.96637312 B10=1.36256839 B11=1.00826248 B12=1.00605494 A1=114.49025526 A2=123.50419607 A3=116.07872726 A4=128.4501019 A5=115.92568971 A6=121.82030618 A7=116.45785973 A8=105.48312518 A9=122.04257434 A10=115.41050172 A11=118.43838067 D1=-0.1413457 D2=0.14838331 D3=-0.16233264 D4=179.90592019 D5=-179.54210511 D6=-179.86991518 D7=-0.06444788 D8=-177.77552501 D9=-166.38608126 D10=-21.4390202 |
B1=1.33118273 B2=1.33838464 B3=1.37597607 B4=1.40790975 B5=1.32454107 B6=1.08407963 B7=1.07985081 B8=1.34388602 B9=0.96634972 B10=1.3608087 B11=1.00545686 B12=1.00317905 A1=114.80329631 A2=123.34621137 A3=116.24728948 A4=128.02712761 A5=116.11714657 A6=121.73410878 A7=116.68547747 A8=106.02627441 A9=122.1371945 A10=116.75246945 A11=119.78971242 D1=-0.1021715 D2=0.07173595 D3=-0.12853005 D4=179.92049474 D5=-179.57017982 D6=-179.8975806 D7=-0.02507294 D8=-178.26608817 D9=-169.50474398 D10=-16.77852188 |
||||
| |
|||||
| |
|||||
| cis | |||||
| C N,1,B1 C,2,B2,1,A1 C,3,B3,2,A2,1,D1,0 C,4,B4,3,A3,2,D2,0 N,1,B5,2,A4,3,D3,0 H,3,B6,2,A5,1,D4,0 H,4,B7,3,A6,2,D5,0 O,1,B8,2,A7,3,D6,0 H,9,B9,1,A8,2,D7,0 N,5,B10,4,A9,3,D8,0 H,11,B11,5,A10,4,D9,0 H,11,B12,5,A11,4,D10,0 |
|||||
| B3P86 | B3LYP | ||||
| B1=1.32506801 B2=1.33677599 B3=1.3787032 B4=1.40372592 B5=1.33179755 B6=1.08780311 B7=1.08278496 B8=1.33886741 B9=0.96579945 B10=1.36434218 B11=1.00819689 B12=1.00638182 A1=114.07672341 A2=124.05051439 A3=116.09791825 A4=128.40119588 A5=115.69883636 A6=121.89664438 A7=115.50866232 A8=106.01649026 A9=122.24639175 A10=115.41069432 A11=117.93873474 D1=-0.13142323 D2=0.19713078 D3=-0.2446351 D4=179.90412971 D5=-179.52121386 D6=-179.96814658 D7=-179.7605362 D8=-177.71030932 D9=-165.27016387 D10=-22.0883644 |
B1=1.32479505 B2=1.33729568 B3=1.37774992 B4=1.40521696 B5=1.33185242 B6=1.08420642 B7=1.0796612 B8=1.34394064 B9=0.96568737 B10=1.36173556 B11=1.00505431 B12=1.00314746 A1=114.38631641 A2=123.89625574 A3=116.29160551 A4=127.99229176 A5=115.88958077 A6=121.76247879 A7=115.69145121 A8=106.55525794 A9=122.39951366 A10=116.98934651 A11=119.54937601 D1=-0.08433136 D2=0.09012435 D3=-0.19063797 D4=179.90899979 D5=-179.57841032 D6=-179.96860067 D7=-179.82752601 D8=-178.27982923 D9=-168.96047155 D10=-16.81122491 |
||||
| |
|||||
| |
||||
| Table 4. Cytosine. Rotational Constants (MHz). Calc on (1) B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimized molecular structure. | ||||
| Calc (1) |
Calc (2) |
Expt. [1] | ||
| trans | A |
3978. |
3975. |
3951.85325(32) |
| B |
2019. |
2011. |
2008.95802(12) |
|
| C |
1340. |
1336. |
1332.47228(8) |
|
| cis | A |
3909. |
3906. |
3889.46510(38) |
| B |
2039. |
2032. |
2026.31804(12) |
|
| C |
1341. |
1337. |
1332.86951(10) |
|