| |
|||||||||
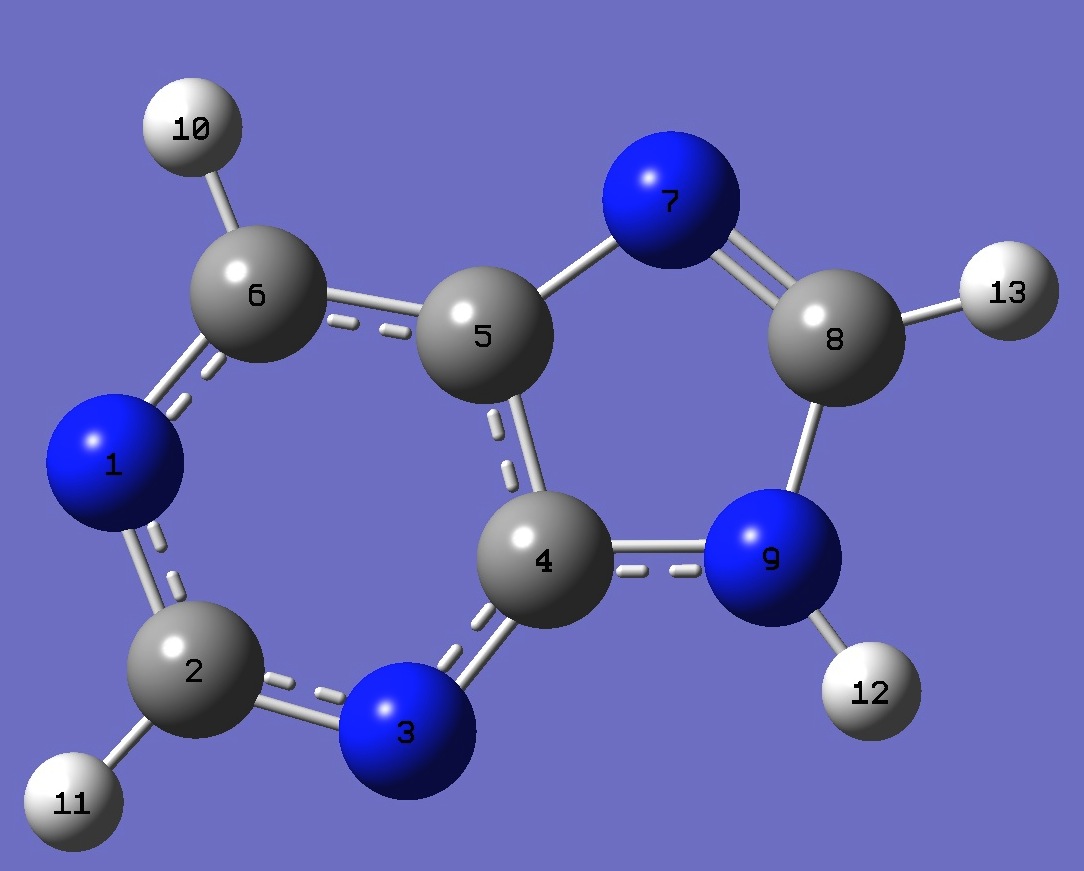
| Table 1. 14N nqcc's in N(9)H-Purine (MHz). Calculation was made on molecular structures given by (1) B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimization. | |||||||||
| |
|||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
|||||||
| N(1) |
Xaa |
- |
3.359 |
- |
3.361 |
- |
3.343(5) |
||
| Xbb | 0.409 |
0.395 |
0.440(6) |
||||||
| Xcc | 2.949 |
2.965 |
2.904(6) |
||||||
| |Xab| | 2.539 |
2.567 |
|||||||
| |
|||||||||
| RMS |
0.033 (1.5 %) |
0.045(2.0 %) |
|||||||
| N(3) |
Xaa | 1.724 |
1.754 |
1.673(7) |
|||||
| Xbb | - |
4.205 |
- |
4.257 |
- |
4.229(9) |
|||
| Xcc | 2.481 |
2.504 |
2.555(9) |
||||||
| |Xab| | 0.070 |
0.062 |
|||||||
| RMS | 0.053 (1.9 %) |
0.058 (2.0 %) |
|||||||
| N(7) |
Xaa | 1.457 |
1.486 |
1.547(7) |
|||||
| Xbb | - |
3.384 |
- |
3.435 |
- |
3.379(9) |
|||
| Xcc | 1.927 |
1.949 |
1.833(9) |
||||||
| |Xab| | 1.684 |
1.677 |
|||||||
| |
|||||||||
| RMS |
0.075 (3.3 %) |
0.082 (3.6 %) |
|||||||
| N(9) | Xaa | 1.423 |
1.460 |
1.489(5) |
|||||
| Xbb | 1.505 |
1.501 |
1.495(7) |
||||||
| Xcc | - |
2.928 |
- |
2.961 |
- |
2.985(7) |
|||
| |Xab| | 0.093 |
0.082 |
|||||||
| |
|||||||||
| RMS |
0.051 (2.6 %) |
0.022 (1.1 %) |
|||||||
| |
|||||||||
| |
|||||||||
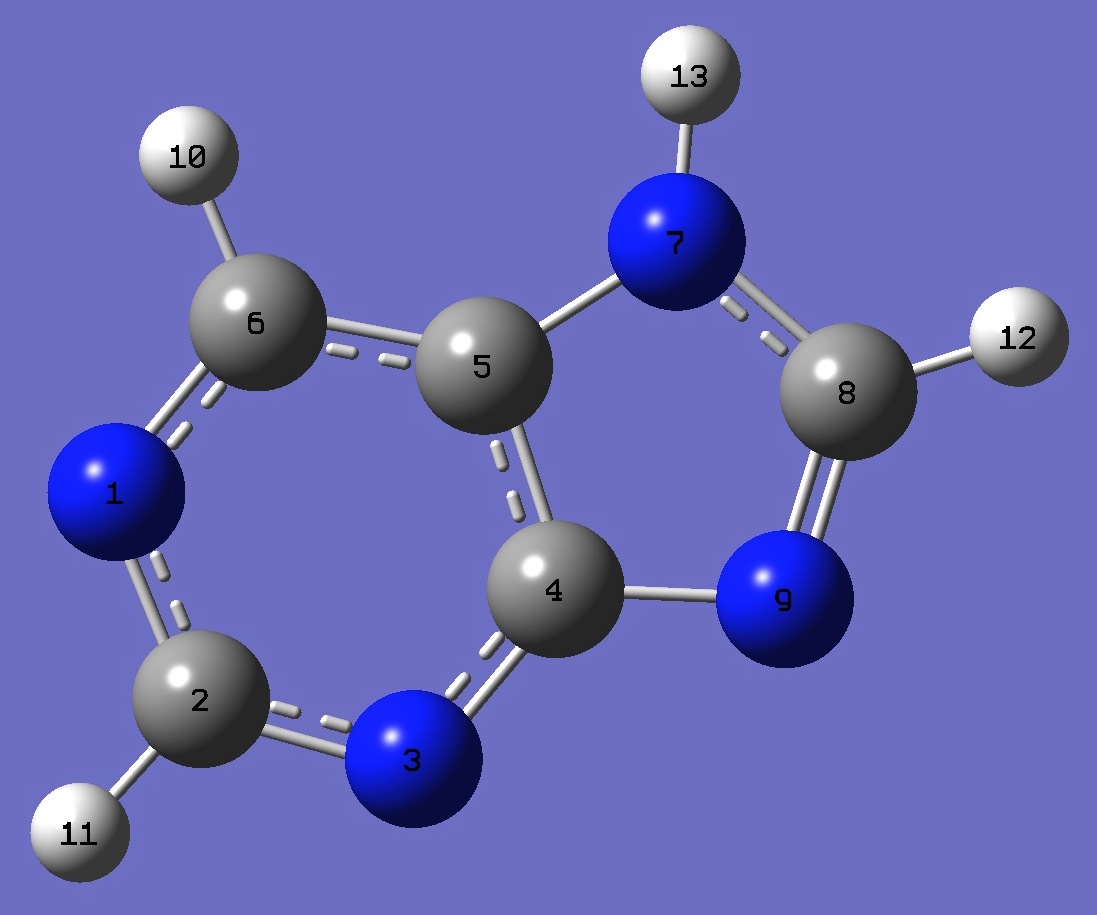
| Table 2. 14N nqcc's in N(7)H-Purine (MHz). Calculation was made on molecular structures given by (1) B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimization. | |||||||||
| |
|||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
|||||||
| N(1) |
Xaa |
- |
3.394 |
- |
3.394 |
- |
3.560(15) | ||
| Xbb | 0.406 |
0.386 |
0.566(17) |
||||||
| Xcc | 2.988 |
3.008 |
2.994(17) |
||||||
| |Xab| | 2.482 |
2.512 |
|||||||
| |
|||||||||
| RMS |
0.133 (5.6 %) |
0.142 (6.0 %) |
|||||||
| N(3) |
Xaa | 1.498 |
1.525 |
0.695(11) |
|||||
| Xbb | - |
4.547 |
- |
4.592 |
- |
2.744(20) |
|||
| Xcc | 3.049 |
3.067 |
2.049(20) |
||||||
| |Xab| | 0.053 |
0.051 |
|||||||
| RMS | 1.278 (70. %) |
1.309 (72. %) |
|||||||
| N(7) |
Xaa | 1.633 |
1.660 |
1.254(11) | |||||
| Xbb | 1.516 |
1.511 |
1.532(16) | ||||||
| Xcc | - |
3.149 |
- |
3.171 |
- |
2.786(16) |
|||
| |Xab| | 0.104 |
0.094 |
|||||||
| |
|||||||||
| RMS |
0.303 (16. %) | 0.323 (17. %) | |||||||
| N(9) | Xaa | 1.414 |
1.448 |
1.541(11) |
|||||
| Xbb | - |
3.417 |
- |
3.460 |
- |
3.239(18) |
|||
| Xcc | 2.002 |
2.011 |
1.699(18) |
||||||
| |Xab| | 1.593 |
1.591 |
|||||||
| |
|||||||||
| RMS |
0.216 (10. %) |
0.227 (11. %) |
|||||||
| |
|||||||||
| |
||||
| Table 4. Purine. Rotational Constants (MHz). Calc on (1) B3P86/6-31G(3d,3p) and (2) B3LYP/cc-pVTZ optimized molecular structure. | ||||
| Calc (1) |
Calc (2) |
Expt. [1] | ||
| N(9)H | A |
4157. |
4163. |
4125.8895(2) |
| B |
1765. |
1760. |
1755.1720(1) |
|
| C |
1239. |
1237. |
1231.55819(6) |
|
| N(7)H | A |
4160. |
4166. |
4127.3813(3) |
| B |
1761. |
1756. |
1749.7594(3) |
|
| C |
1237. |
1235. |
1229.42760(7) |
|