|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
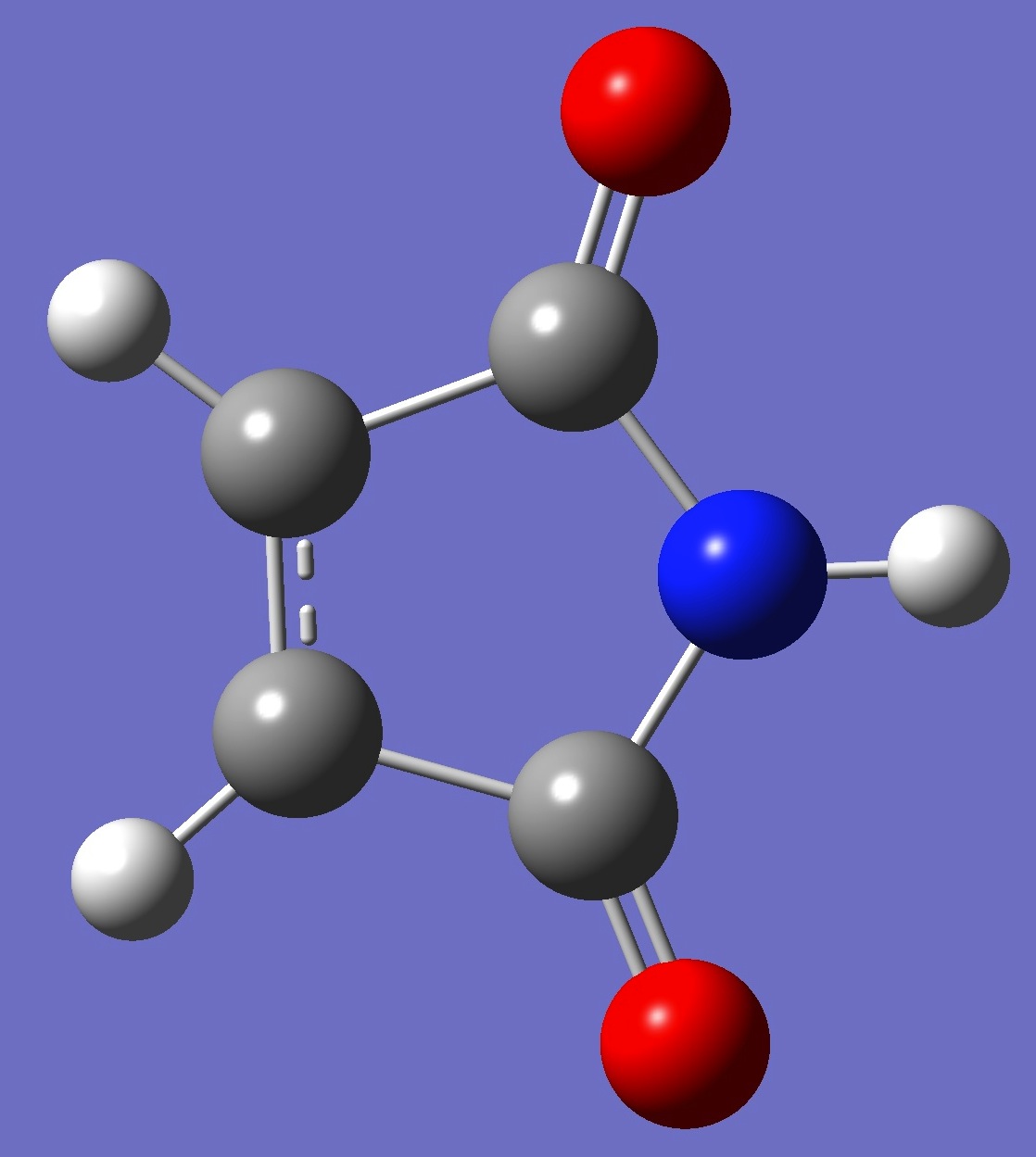
C4H3NO2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen |
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
|
in Maleimide
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen nqcc's in maleimide,
as well as a best-fit molcular structure, were determined by Pejlovas
et al. [1]. Calculation of the nqcc's was made here on this
best-fit
structure, and on an ropt structure given by B3LYP/cc-pVTZ optimization. These are compared with the experimental nqcc's in
Table 1. Structure parameters are given in Z-matrix format in Table 2.
|
|
|
|
|
|
|
|
|
|
|
|
|
In Table 1, subscripts a,b,c
refer to the
principal axes of the inertia tensor. RMS is the root mean square
difference between calculated and experimental diagonal nqcc's
(percentage of the average of the magnitudes of the experimental
nqcc's). RSD is the calibration residual standard deviation for
the B3PW91/6-311+G(df,pd) model for calculation of the nitrogen
efg's/nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
Table 1. Nitrogen nqcc's
in Maleimide (MHz). Calculation was made on best-fit and ropt structures.
|
|
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc /best-fit |
|
Calc /ropt |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.598
|
|
1.588
|
|
1.6151(35) *
|
|
|
|
Xbb |
|
1.948
|
|
1.924
|
|
1.9282(35) *
|
|
|
|
Xcc |
-
|
3.536
|
-
|
3.512
|
-
|
3.5434(35) *
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.012 (0.53 %)
|
|
0.024 (1.0 %)
|
|
|
|
|
|
RSD |
|
0.030 (1.3 %)
|
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* Calculated here from 1.5Xaa = 2.4227(53) and 0.25(Xbb - Xcc) = 1.3679(15) MHz using Kisiel's QDIAG program.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
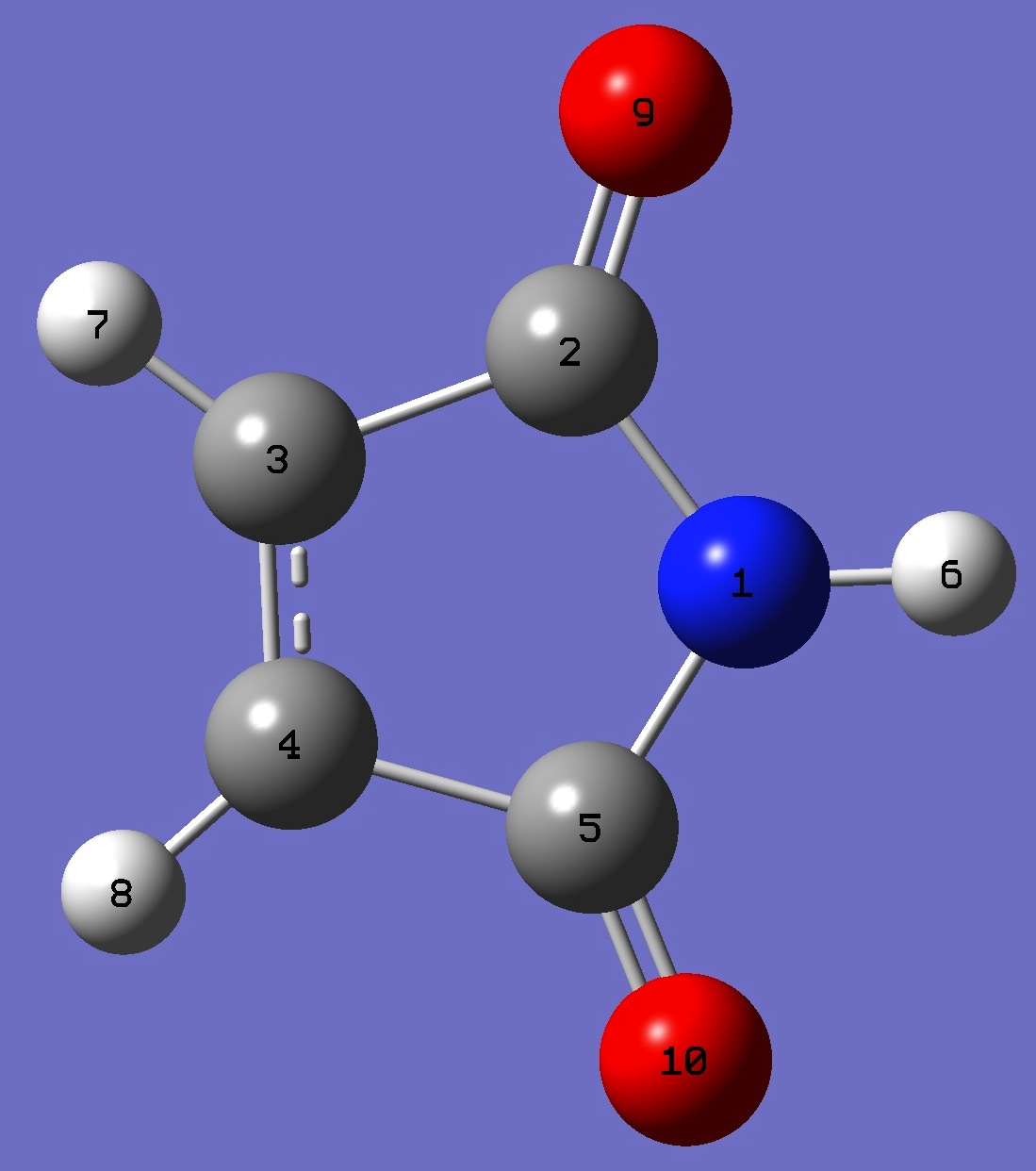
Table 2. Maleimide.
Molecular structure parameters best-fit [1] and ropt = B3LYP/cc-pVTZ optimization (Å and degrees). Best-fit parameters derived from a,b,c coordinates given in Ref. [1].
|
|
|
|
|
|
N
C 1 B1
C 2 B2 1 A1
C 3 B3 2 A2 1 D1
C 1 B4 2 A3 3 D2
H 1 B5 5 A4 4 D3
H 3 B6 2 A5 1 D4
H 4 B7 3 A6 2 D5
O 2 B8 1 A7 5 D6
O 5 B9 1 A8 2 D7
|
|
|
|
|
|
|
|
|
|
Best-Fit
|
ropt |
|
|
|
|
|
|
B1 1.38817949
B2 1.49802181
B3 1.37400010
B4 1.38817949
B5 1.00700007
B6 1.07875585
B7 1.07875585
B8 1.20597397
B9 1.20597397
A1 106.17327903
A2 107.96324007
A3 111.72696181
A4 124.13651910
A5 122.75482704
A6 129.28193289
A7 126.39182142
A8 126.39182142
D1 0.00000000
D2 -0.00000000
D3 180.00000000
D4 180.00000000
D5 180.00000000
D6 180.00000000
D7 -180.00000000
|
B1 1.39295086
B2 1.50074709
B3 1.33018629
B4 1.39295086
B5 1.00652807
B6 1.07799513
B7 1.07799513
B8 1.20449212
B9 1.20449212
A1 105.14507453
A2 108.97771454
A3 111.75442184
A4 124.12278908
A5 121.76902325
A6 129.25326220
A7 126.40817071
A8 126.40817071
D1 0.00000000
D2 -0.00000000
D3 180.00000000
D4 180.00000000
D5 180.00000000
D6 180.00000000
D7 -180.00000000
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] A.M.Pejlovas, O.Oncer, L.Kang, and S.G.Kukolich, J.Mol.Spectrosc. 319,26(2016).
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Pyrrole
|
Hydantoin
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Maleimide.html |
|
|
|
|
|
|
Last
Modified 12 Dec 2015 |
|
|
|
|
|
|
|
|
|
|

