|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Quinoline |
PDF |
|
|
|
|
|
|
and |
|
|
|
|
|
|
|
|
|
Isoquinoline |
PDF |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
in Quinoline and Isoquinoline
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen nqcc's in quinoline and isoquinoline were determined by Kisiel et al. [1]. |
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the nqcc's was made here on molecular structures given by B3P86/6-31G(d,p) and B3P86/6-31G(3d,3p)
optimizations. Calculated and experimental nqcc's are compared in
Tables 1 and 2. Structure parameters are given in Table 3,
rotational constants in Table 4. |
|
|
|
|
|
|
|
|
|
|
|
|
In Tables 1 and 2, subscripts a,b,c refer to the
principal axes of the inertia tensor; x,y,z to the principal axes
of the nqcc tensor.
Ø (degrees) is the angle between its subscripted
parameters. ETA = (Xxx - Xyy)/Xzz. |
|
|
|
|
|
|
|
|
|
|
|
|
RMS is the root mean square
difference between calculated and experimental diagonal nqcc's (percentage of the
average of the magnitudes of the experimental nqcc's). RSD is the
calibration residual standard deviation of the B3PW91/6-311+G(df,pd) model for calculation of nitrogen nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 1. 14N nqcc's
in Quinoline (MHz). Calculation was made
on (1) B3P86/6-31G(d,p) and (2) B3P86/6-31G(3d,3p) optimized structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. (1)
|
|
Calc. (2) |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
|
1.492 |
|
1.482 |
|
1.4629(10) |
|
|
Xbb |
- |
4.695 |
- |
4.702 |
- |
4.6841(11) |
|
|
Xcc |
|
3.204 |
|
3.219 |
|
3.2213(11) |
|
|
|Xab| |
|
0.380 |
|
0.386 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.020 (0.66 %) |
|
0.015 (0.48 %) |
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
1.515 |
|
1.506 |
|
|
|
|
Xyy |
|
3.204 |
|
3.219 |
|
|
|
|
Xzz |
- |
4.718 |
- |
4.726 |
|
|
|
|
ETA |
|
0.358 |
|
0.362 |
|
|
|
|
Øz,a |
|
86.50 |
|
86.44 |
|
|
|
|
Øa,bi |
|
89.77 |
|
89.80 |
|
|
|
|
Øz,bi* |
|
3.27 |
|
3.36 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* Angle between the principal z-axis of the nqcc tensor and the bisector ( bi ) of the CNC angle. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 2. 14N nqcc's
in Isoquinoline (MHz). Calculation was made
on (1) B3P86/6-31G(d,p) and (2) B3P86/6-31G(3d,3p) optimized structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. (1)
|
|
Calc. (2) |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
- |
3.478 |
- |
3.486 |
- |
3.5114(15) |
|
|
Xbb |
|
0.109 |
|
0.104 |
|
0.1304(19) |
|
|
Xcc |
|
3.369 |
|
3.382 |
|
3.3810(19) |
|
|
|Xab| |
|
2.629 |
|
2.620 |
|
2.81(42) |
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
0.024 (1.0 %) |
|
0.021 (0.89 %) |
|
|
|
|
RSD |
|
0.030 (1.3 %) |
|
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
1.498 |
|
1.485 |
|
|
|
|
Xyy |
|
3.369 |
|
3.382 |
|
|
|
|
Xzz |
- |
4.867 |
- |
4.868 |
- |
5.04(25) |
|
|
ETA |
|
0.384 |
|
0.390 |
|
|
|
|
Øz,a |
|
27.85 |
|
27.79 |
|
28.5(20) |
|
|
Øa,bi |
|
29.96 |
|
29.95 |
|
|
|
|
Øz,bi* |
|
2.11 |
|
2.16 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* Angle between the principal z-axis of the nqcc tensor and the bisector ( bi ) of the CNC angle. |
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
| Table 3. NC bond
lengths and CNC angle (Å and degrees). Complete
B3P86/6-31G(3d,3p) structure parameters are given here in Z-matrix format. |
| |
|
|
|
|
|
|
ropt (1) B3P86/6-31G(d,p) |
|
|
ropt (2) B3P86/6-31G(3d,3p) |
| |
|
|
|
|
| Quinoline |
|
ropt (1) |
ropt (2) |
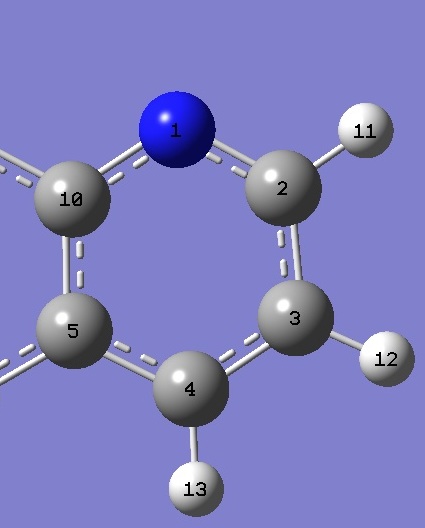 |
|
|
|
| C(10)N |
1.3612 |
1.3604 |
| NC(2) |
1.3145 |
1.3122 |
| C(10)NC(2) |
117.61 |
117.60 |
| |
|
|
| |
|
|
|
|
| Isoquinoline |
|
|
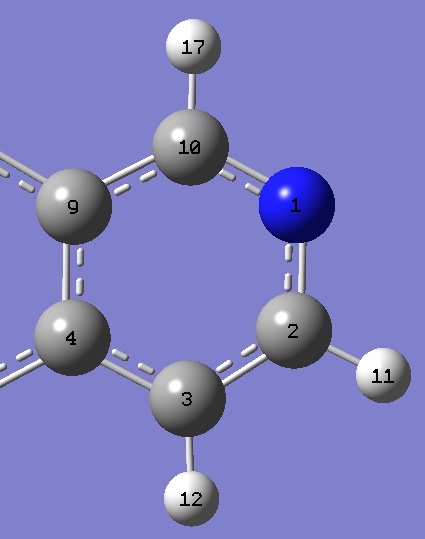 |
|
|
|
| C(10)N |
1.3132 |
1.3113 |
| NC(2) |
1.3580 |
1.3565 |
| C(10)NC(2) |
117.24 |
117.18 |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
| Table 4. Rotational Constants (MHz). Normal Species. |
| |
|
|
|
|
|
ropt (1) = B3P86/6-31G(d,p) optimization. |
|
ropt (2) = B3P86/6-31G(3d,3p) optimization. |
| |
|
|
|
|
|
Quinoline |
|
|
|
|
|
|
|
|
|
Calc. ropt (1) |
Calc. ropt (2) |
Expt. |
|
|
|
|
|
|
A |
3160.0 |
3168.0 |
3145.533013(79)
|
|
B |
1275.8 |
1279.4 |
1271.577972(74)
|
|
C |
908.8 |
911.4 |
905.739406(44)
|
|
|
|
|
|
|
Isoquinoline |
|
|
|
|
|
|
|
|
A |
3213.4 |
3223.1 |
3199.00020(30) |
|
B |
1242.1 |
1246.0 |
1237.931586(74) |
|
C |
895.8 |
898.6 |
892.753595(48) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1] Z.Kisiel, O.Desyatnyk, L.Pszczółkowski, S.B.Charnley, and P.Ehrenfreund, J.Mol.Spectrosc. 217,115(2003). |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Pyridine |
Pyrimidine |
Pyrazine |
1,3,5-Triazine
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Quinoline.html |
|
|
|
|
|
|
Last
Modified 16 Jan 2006 |
|
|
|
|
|
|
|
|
|
|

