| |
||||||||
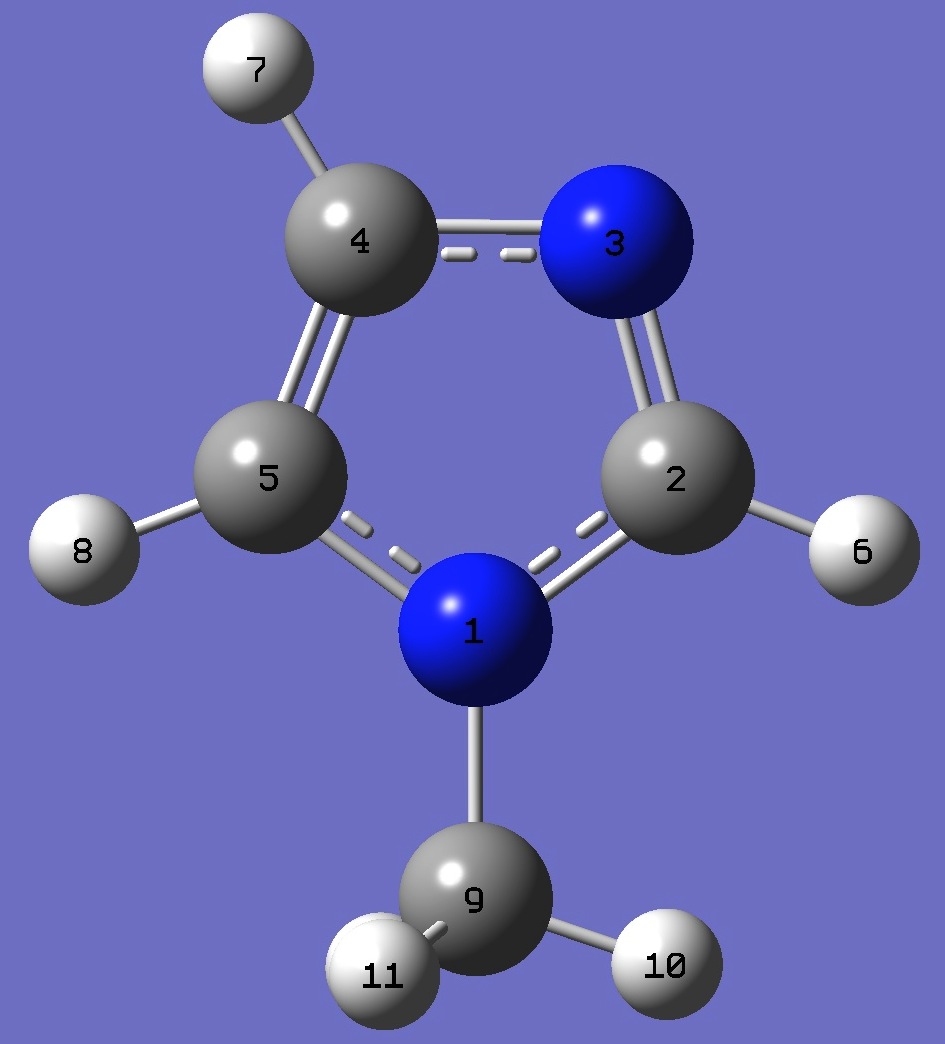
| Table 1. Pyrrolic 14N(1) nqcc's in N-Methylimidazole (MHz). Calculation was made
on the ropt = B3LYP/cc-pVTZ optimized structure, and on the heavy atom rs structure. |
||||||||
| |
||||||||
| Calc /ropt | Calc /rs | Expt [1] |
||||||
| |
||||||||
| Xaa | 1.720 |
1.639 |
1.7347(51) |
|||||
| Xbb - Xcc | 3.535 |
3.186 |
3.6373(83) |
|||||
| Xbb | 0.908 |
0.773 |
0.9513 * |
|||||
| Xcc | - |
2.628 |
- |
2.412 |
- |
2.6860 * |
||
| Xab | - |
0.079 |
- |
0.076 |
||||
| RMS |
0.043 (2.4 %) |
0.196 (11. %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| |
||||||||
| Xxx | 0.900 |
0.767 |
||||||
| Xyy | 1.728 |
1.645 |
||||||
| Xzz | - |
2.628 |
- |
2.412 |
||||
| ETA |
0.315 |
0.364 |
||||||
| ěz,c |
zero |
zero |
||||||
| |
||||||||
| |
||||||||
| Table 2. Pyridinic 14N(3) nqcc's in N-Methylimidazole (MHz). Calculation was made on the ropt = B3LYP/cc-pVTZ optimized structure, and on the heavy atom rs structure. | ||||||||
| |
||||||||
| Calc /ropt | Calc /rs | Expt [1] |
||||||
| |
||||||||
| Xaa | - |
2.106 |
- |
1.947 |
- |
2.0837(52) |
||
| Xbb - Xcc | - |
2.271 |
- |
2.154 |
- |
2.200(10) |
||
| Xbb | - |
0.083 |
- |
0.104 |
- |
0.0582 * |
||
| Xcc | 2.189 |
2.050 |
2.1419 * |
|||||
| Xab | - |
2.770 |
- |
2.012 |
||||
| RMS |
0.033 (2.3 %) |
0.083 (5.8 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| |
||||||||
| Xxx | 1.855 |
1.933 |
||||||
| Xyy | 2.189 | 2.050 |
||||||
| Xzz | - |
4.044 |
- |
3.984 |
||||
| ETA |
0.0826 |
0.0294 |
||||||
| ěz,bi** | ||||||||
| |
||||||||
| Table 4. N-Methylimidazole: Rotational Constants (MHz). |
|||||
| |
|||||
| Calc /ropt | Calc /rs | Expt [1] | |||
| |
|||||
| A |
9226. |
9205. |
9156.7503(50) | ||
| B |
3621. |
3622. |
3609.9582(16) | ||
| C |
2644. |
2642 |
2632.4462(16) | ||