| |
||||||||
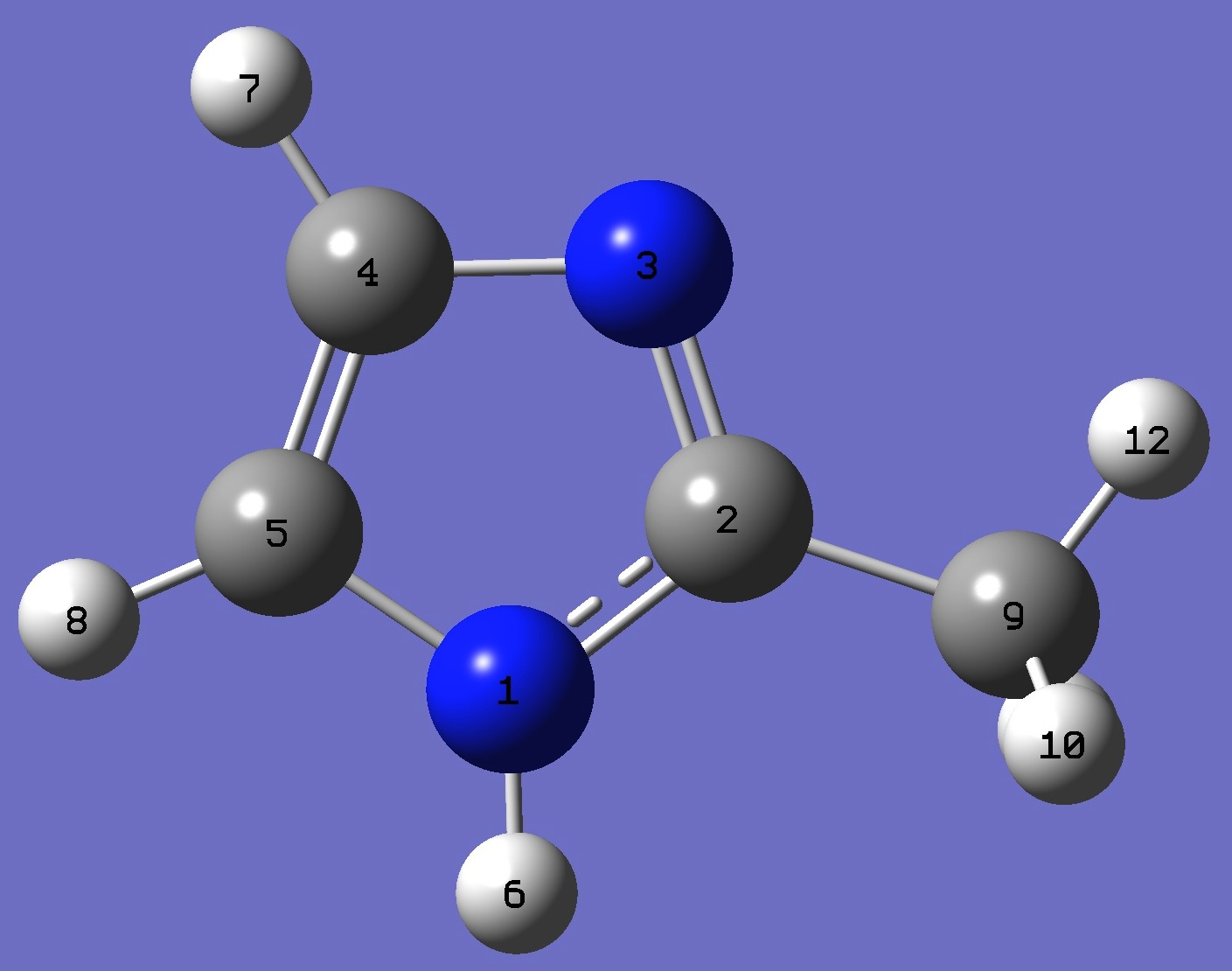
| Table 1. Pyrrolic 14N(1) nqcc's in 2-Methylimidazole (MHz). Calculation was made
on the ropt = B3LYP/cc-pVTZ optimized structure, and on the heavy atom rs structure. |
||||||||
| |
||||||||
| Calc /ropt | Calc /rs | Expt [1] |
||||||
| |
||||||||
| Xaa | 1.266 |
1.276 |
1.3010(59) |
|||||
| Xbb - Xcc | 4.236 |
4.082 |
4.178(10) |
|||||
| Xbb | 1.485 |
1.403 |
1.4385 * |
|||||
| Xcc | - |
2.751 |
- |
2.679 |
- |
2.7395 * |
||
| Xab | 0.132 |
0.258 |
||||||
| RMS |
0.025 (1.4 %) |
0.043 (2.4 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| |
||||||||
| Xxx | 1.204 |
1.074 |
||||||
| Xyy | 1.547 |
1.605 |
||||||
| Xzz | - |
2.751 |
- |
2.679 |
||||
| ETA |
0.125 |
0.198 |
||||||
| ěz,c |
zero |
zero |
||||||
| |
||||||||
| |
||||||||
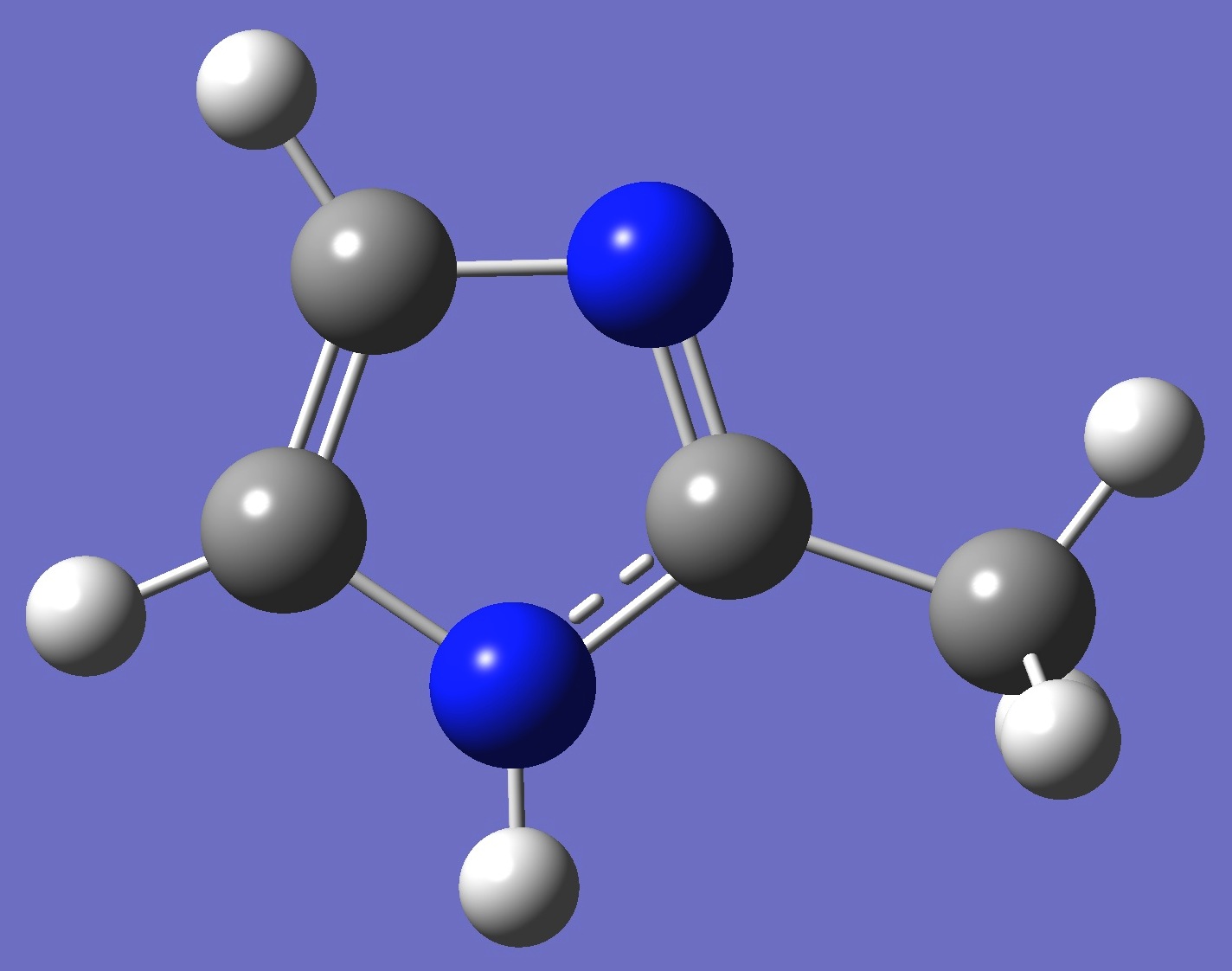
| Table 2. Pyridinic 14N(3) nqcc's in 4-Methylimidazole (MHz). Calculation was made on the ropt = B3LYP/cc-pVTZ optimized structure, and on the heavy atom rs structure. | ||||||||
| |
||||||||
| Calc /ropt | Calc /rs | Expt [1] |
||||||
| |
||||||||
| Xaa | 1.374 |
1.455 |
1.3742(64) |
|||||
| Xbb - Xcc | - |
5.463 |
- |
4.591 |
- |
5.399(10) |
||
| Xbb | - |
3.418 |
- |
3.023 |
- |
3.3866 * |
||
| Xcc | 2.044 |
1.568 |
2.0124 * |
|||||
| Xab | 1.703 |
1.691 |
||||||
| RMS |
0.026 (1.3 %) |
0.157 (7.7 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| |
||||||||
| Xxx | 1.918 |
1.816 |
||||||
| Xyy | 2.044 | 2.198 |
||||||
| Xzz | - |
3.962 |
- |
4.014 |
||||
| ETA |
0.0319 |
0.0949 |
||||||
| ěz,bi** | ||||||||
| |
||||||||
| Table 4. 2-Methylimidazole: Rotational Constants (MHz). |
|||||
| |
|||||
| Calc /ropt | Calc /rs | Expt [1] | |||
| |
|||||
| A |
8979. |
8910. |
8892.9063(26) | ||
| B |
3605. |
3614. |
3601.19596(95) | ||
| C |
2614. |
2613. |
2604.8287(98) | ||