| |
||||||||
| Table
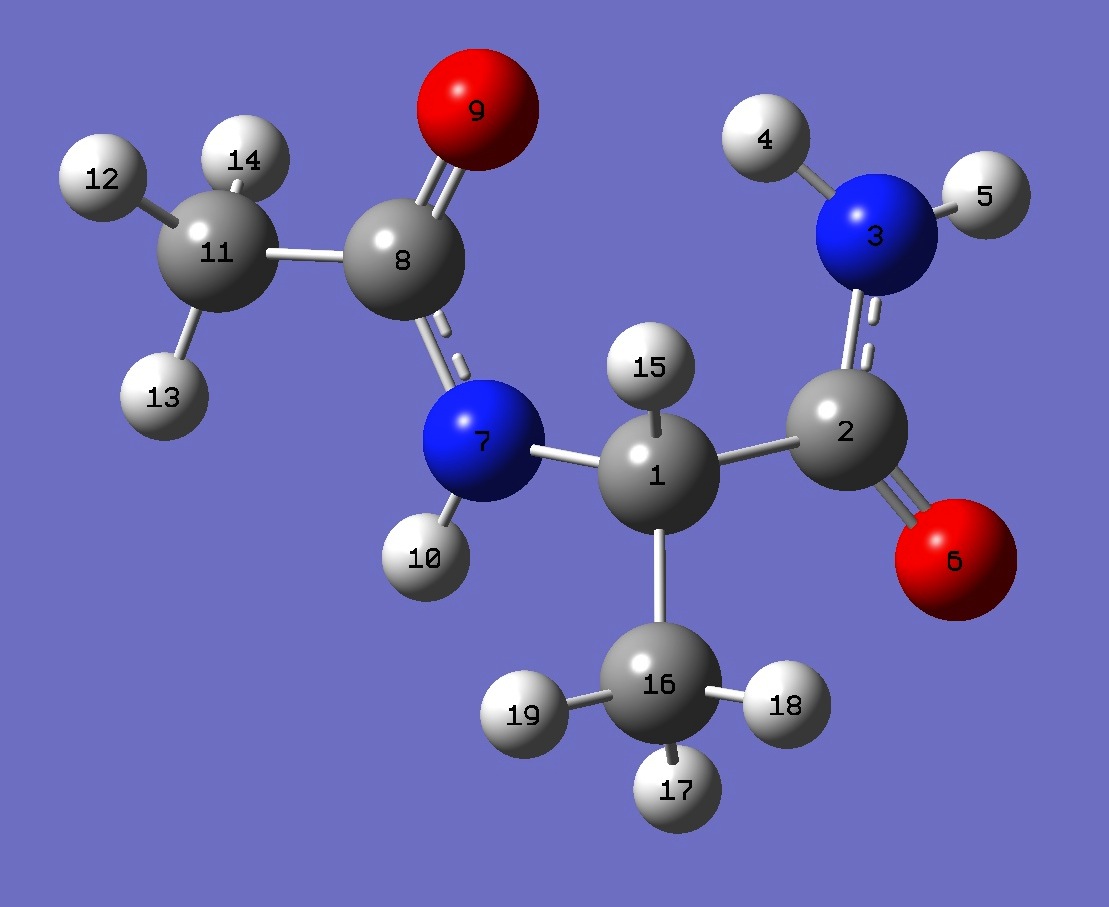
1. 14N nqcc's in C7eq N-Acetyl-Alaninamide (MHz).
Calculation was made on (1) mPW1PW91/6-31G(3d,3p) and (2)
mPW1PW91/cc-pVDZ optimized structures. Atomic numbering is shown
above. |
||||||||
| |
||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
||||||
| Xaa N(7) | 1.521 |
1.511 |
1.5731(33) |
|||||
| Xbb | - |
0.896 |
- |
0.917 |
- |
0.9751(70) |
||
| Xcc | - |
0.625 |
- |
0.594 |
- |
0.5979(70) |
||
| Xab | 1.056 |
1.071 |
||||||
| Xac | - |
1.325 |
- |
1.310 |
||||
| Xbc | 2.642 |
2.657 |
||||||
| |
||||||||
| RMS |
0.057 (5.4 %) |
0.049 (4.7 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xaa N(3) | 0.168 |
0.102 |
0.1133(88) |
|||||
| Xbb | 2.082 |
2.089 |
2.0961(76) |
|||||
| Xcc | - |
2.250 |
- |
2.191 |
- |
2.2095(76) |
||
| Xab | 0.315 |
0.322 |
||||||
| Xac | 2.125 |
2.139 |
||||||
| Xbc | - |
0.618 |
- |
0.616 |
||||
| RMS |
0.040 (2.7 %) |
0.015 (1.0 %) | ||||||
| RSD |
0.030 (1.3 %) | 0.030 (1.3 %) | ||||||
| |
||||||||
| |
||||||||
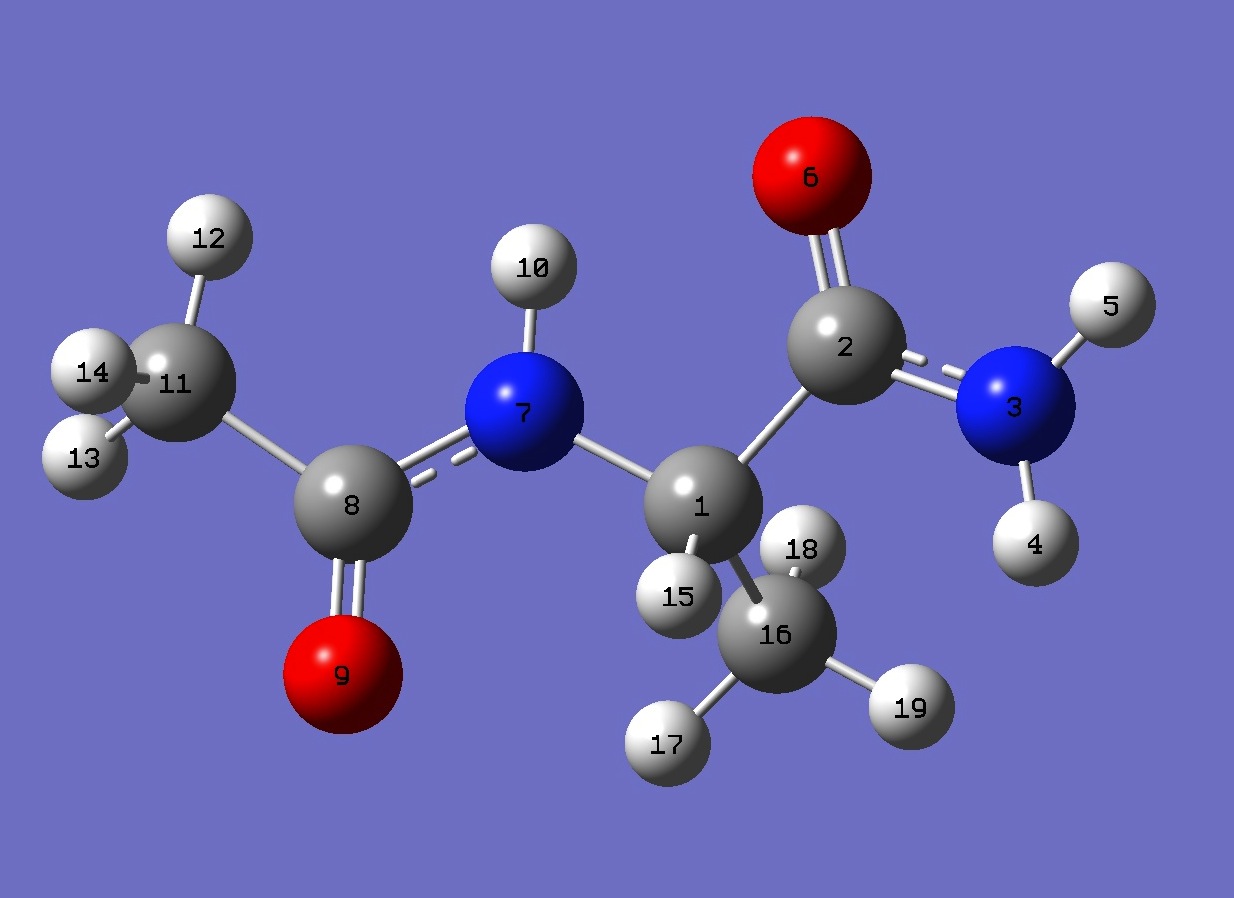
| Table 2. 14N nqcc's in C5 N-Acetyl-Alaninamide (MHz).
Calculation was made on (1) mPW1PW91/6-31G(3d,3p) and (2)
mPW1PW91/cc-pVDZ optimized structures. Atomic numbering is shown
above. |
||||||||
| |
||||||||
| Calc (1) |
Calc (2) |
Expt. [1] |
||||||
| Xaa N(7) | 2.095 |
2.114 |
2.085(25) |
|||||
| Xbb | 1.159 |
1.109 |
1.232(48) |
|||||
| Xcc | - |
3.254 |
- |
3.223 |
- |
3.316(48) |
||
| Xab | 0.598 |
0.579 |
||||||
| Xac | 0.415 |
0.370 |
||||||
| Xbc | - |
1.755 |
- |
1.784 |
||||
| |
||||||||
| RMS |
0.055 (2.5 %) |
0.091 (4.1 %) |
||||||
| RSD | 0.030 (1.3 %) |
0.030 (1.3 %) | ||||||
| Xaa N(3) | 1.593 |
1.627 |
1.754(21) |
|||||
| Xbb | 0.965 |
1.163 |
0.998(37) |
|||||
| Xcc | - |
2.558 |
- |
2.790 |
- |
2.752(37 |
||
| Xab | - |
0.619 |
- |
0.547 |
||||
| Xac | - |
1.466 |
- |
1.469 |
||||
| Xbc | - |
2.206 |
- |
2.053 |
||||
| RMS |
0.147 (8.0 %) |
0.122 (6.7 %) |
||||||
| RSD |
0.030 (1.3 %) | 0.030 (1.3 %) | ||||||
| |
||||||||
| Table 3. N-Acetyl-Alaninamide, mPW1PW91/6-31G(3d,3p) and mPW1PW91/cc-pVDZ optimized molecular structures
(Å and degrees). Atomic numbering is given in Fig. 1. |
|||
| C C,1,B1 N,2,B2,1,A1 H,3,B3,2,A2,1,D1,0 H,3,B4,2,A3,1,D2,0 O,2,B5,1,A4,3,D3,0 N,1,B6,2,A5,6,D4,0 C,7,B7,1,A6,2,D5,0 O,8,B8,7,A7,1,D6,0 H,7,B9,1,A8,2,D7,0 C,8,B10,7,A9,1,D8,0 H,11,B11,8,A10,7,D9,0 H,11,B12,8,A11,7,D10,0 H,11,B13,8,A12,7,D11,0 H,1,B14,7,A13,8,D12,0 C,1,B15,7,A14,8,D13,0 H,16,B16,1,A15,7,D14,0 H,16,B17,1,A16,7,D15,0 H,16,B18,1,A17,7,D16,0 |
|||
| _________ C7eq
_________
|
___________ C5 ___________ | ||
| 6-31(3d,3p) |
cc-pVDZ |
6-31(3d,3p) | cc-pVDZ |
| B1=1.54133991 B2=1.3504847 B3=1.01352472 B4=1.00609265 B5=1.2158468 B6=1.45682713 B7=1.35145349 B8=1.22586494 B9=1.00638072 B10=1.50798754 B11=1.0908549 B12=1.09077484 B13=1.09085972 B14=1.09423293 B15=1.51464039 B16=1.09189443 B17=1.08960657 B18=1.09166972 A1=113.56395768 A2=119.43127541 A3=117.85012223 A4=122.22244052 A5=110.3291126 A6=123.1582746 A7=122.18613026 A8=117.64013735 A9=116.33354559 A10=108.47582069 A11=113.68576213 A12=108.77638759 A13=106.23467508 A14=110.20620893 A15=110.73157694 A16=110.13032798 A17=110.47371312 D1=-9.98305474 D2=-172.69055665 D3=179.58320319 D4=-108.94205182 D5=-82.75038347 D6=2.11332549 D7=94.65701064 D8=-177.47663757 D9=122.18703553 D10=0.63442858 D11=-121.27833965 D12=33.98119669 D13=154.08113944 D14=63.98048361 D15=-177.97524045 D16=-57.66720585 |
B1=1.54237922 B2=1.3534554 B3=1.01912826 B4=1.01118202 B5=1.21841506 B6=1.45850048 B7=1.35470236 B8=1.22864561 B9=1.0108554 B10=1.50860541 B11=1.0978958 B12=1.09737688 B13=1.09833483 B14=1.10065205 B15=1.51462244 B16=1.09883862 B17=1.09685547 B18=1.09846068 A1=113.21889542 A2=118.92868902 A3=117.67463849 A4=122.29628261 A5=110.32351133 A6=122.81554882 A7=122.19286157 A8=117.77550451 A9=116.17039492 A10=108.36054122 A11=113.90393121 A12=108.73684203 A13=105.90773026 A14=110.37097149 A15=110.54106589 A16=110.1450115 A17=110.62600084 D1=-11.30594096 D2=-171.79352363 D3=179.59319238 D4=-107.46192743 D5=-82.06693251 D6=1.96323544 D7=93.5866864 D8=-177.67566378 D9=124.42347501 D10=2.6583047 D11=-119.46308379 D12=34.44367697 D13=154.67307019 D14=66.19665174 D15=-176.03491241 D16=-55.72032775 |
B1=1.52587222 B2=1.35250642 B3=1.00385139 B4=1.0054964 B5=1.21759447 B6=1.43953491 B7=1.35359736 B8=1.22084772 B9=1.01009652 B10=1.51023163 B11=1.09077594 B12=1.09090586 B13=1.09089513 B14=1.09633283 B15=1.53138596 B16=1.08999782 B17=1.09190211 B18=1.09217387 A1=115.44229387 A2=121.82937869 A3=118.22194107 A4=121.85511958 A5=106.9332641 A6=121.70682308 A7=122.14949916 A8=115.43786806 A9=115.84978554 A10=113.52957421 A11=108.70142368 A12=108.6512826 A13=108.28406725 A14=112.19960667 A15=108.88157994 A16=110.34616132 A17=111.35481625 D1=-10.24145238 D2=-178.63868374 D3=177.89416034 D4=-16.09247542 D5=-158.07533317 D6=-2.94330961 D7=14.03336496 D8=177.20752411 D9=-0.28348111 D10=121.39211168 D11=-122.03233539 D12=-39.88763858 D13=79.94963013 D14=-59.20975083 D15=60.52812251 D16=-178.92621616 |
B1=1.52846614 B2=1.35461876 B3=1.00844247 B4=1.01056502 B5=1.22056961 B6=1.43967675 B7=1.35819521 B8=1.22271008 B9=1.01531037 B10=1.51130952 B11=1.09737465 B12=1.0975642 B13=1.09884585 B14=1.10370578 B15=1.53106778 B16=1.09724535 B17=1.09910058 B18=1.09936061 A1=115.36668079 A2=121.83472705 A3=118.04144208 A4=121.92854082 A5=106.76980628 A6=121.34013781 A7=122.10773893 A8=114.61203179 A9=115.51573534 A10=113.61545672 A11=108.65998156 A12=108.66669936 A13=108.41990492 A14=112.14748206 A15=108.64722298 A16=110.44135935 A17=111.71445677 D1=-10.32506838 D2=-177.27657943 D3=178.30419082 D4=-10.7768944 D5=-155.15125291 D6=-6.1043399 D7=10.6037143 D8=174.08182087 D9=6.20651782 D10=128.26093653 D11=-115.4776002 D12=-37.44824413 D13=82.07542424 D14=-59.11952169 D15=60.47094936 D16=-178.69765853 |
| |
|||
| |
||||
| Table 4. N-Acetyl-Alaninamide, mPW1PW91/6-31G(3d,3p), mPW1PW91/cc-pVDZ, and experimental rotational constants (MHz). |
||||
| 6-31G(3d,3p) |
cc-pVDZ |
Expt. [1] | ||
| C7eq | A |
2607. |
2591. |
2598.76659(80) |
| B |
1132. |
1134. |
1125.71115(18) |
|
| C |
917. |
919. |
914.49488(20) |
|
| C5 | A |
3082. |
3051. |
3100.286(32) |
| B |
959. |
966. |
955.11278(23) |
|
| C |
817. |
814. |
812.74871(20) |
|