|
|
|
|
|
|
|
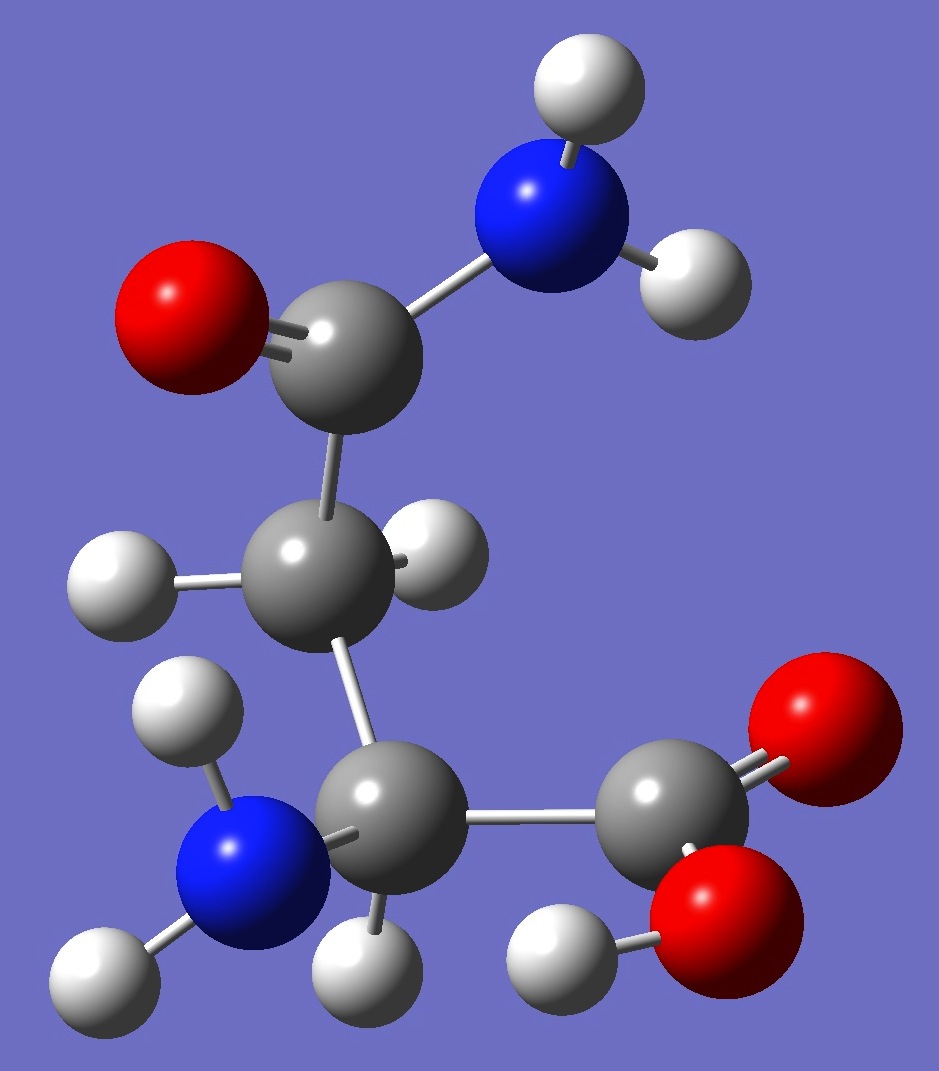
Table 3. Asparagine. Optimized structure parameters, mPW1PW91/6-31G(3d,3p) and mPW1PW91/cc-pVDZ (Å and
degrees).
|
| |
|
|
|
|
|
| 
|
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
O,1,B4,2,A3,3,D2,0
N,1,B5,5,A4,2,D3,0
H,6,B6,1,A5,5,D4,0
H,6,B7,1,A6,5,D5,0
N,3,B8,2,A7,1,D6,0
H,9,B9,3,A8,2,D7,0
H,9,B10,3,A9,2,D8,0
O,4,B11,3,A10,2,D9,0
O,4,B12,3,A11,2,D10,0
H,13,B13,4,A12,3,D11,0
H,3,B14,2,A13,1,D12,0
H,2,B15,1,A14,5,D13,0
H,2,B16,1,A15,5,D14,0
|
|
|
|
|
|
|
|
6-31G(3d,3p) |
|
cc-pVDT
|
|
|
|
B1=1.51475314
B2=1.53500721
B3=1.53717068
B4=1.2223499
B5=1.35302474
B6=1.00787196
B7=1.00737634
B8=1.45999395
B9=1.01860543
B10=1.01085982
B11=1.20564967
B12=1.32185771
B13=0.98804346
B14=1.0947437
B15=1.09037988
B16=1.09088692
A1=109.77175124
A2=110.20550098
A3=120.97942646
A4=123.07480179
A5=119.13893368
A6=116.89592093
A7=115.48791673
A8=109.55850052
A9=111.43634111
A10=122.88406843
A11=113.40970674
A12=103.68951685
A13=108.12347368
A14=111.94654371
A15=107.70105107
D1=-66.8218858
D2=-76.70650846
D3=-178.63267546
D4=161.44505129
D5=9.36839735
D6=57.27251815
D7=-25.51052999
D8=93.22267672
D9=-37.42046057
D10=143.49889332
D11=-5.43176203
D12=179.20061005
D13=162.23080348
D14=42.20738525
|
|
B1=1.5161851
B2=1.53467115
B3=1.53895678
B4=1.22532909
B5=1.3560274
B6=1.01333004
B7=1.01303828
B8=1.46074444
B9=1.02561514
B10=1.01711859
B11=1.208893
B12=1.32339334
B13=0.99483808
B14=1.10239598
B15=1.09661474
B16=1.0979569
A1=109.5837353
A2=110.21329436
A3=121.12227224
A4=123.22028336
A5=118.18474617
A6=116.26832121
A7=115.79762268
A8=108.53984533
A9=111.00579933
A10=123.07787633
A11=112.90537783
A12=102.72480126
A13=108.30781239
A14=111.9935966
A15=107.8092016
D1=-65.79917172
D2=-73.71006166
D3=-178.50406986
D4=158.02425372
D5=10.44525698
D6=57.82011142
D7=-28.05647717
D8=88.66570601
D9=-34.33765225
D10=146.42054528
D11=-6.62020622
D12=179.90871991
D13=165.32099128
D14=45.35725641
|
|
|
|
|
|
|
|
|