|
| |
|
|
|
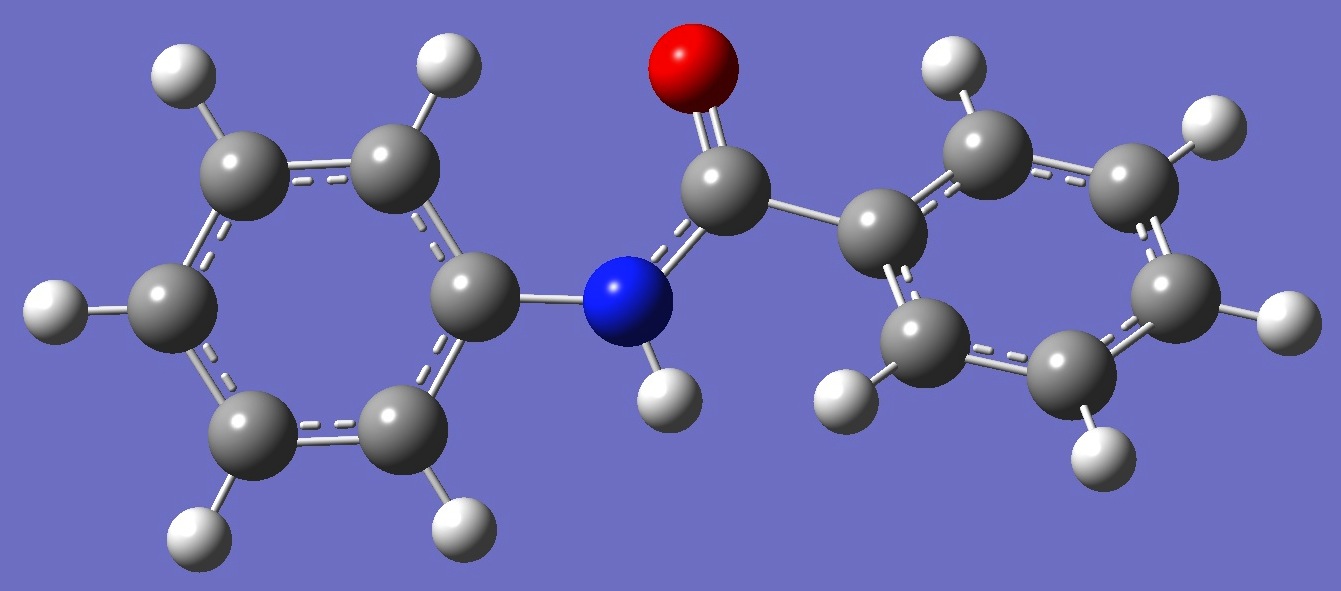
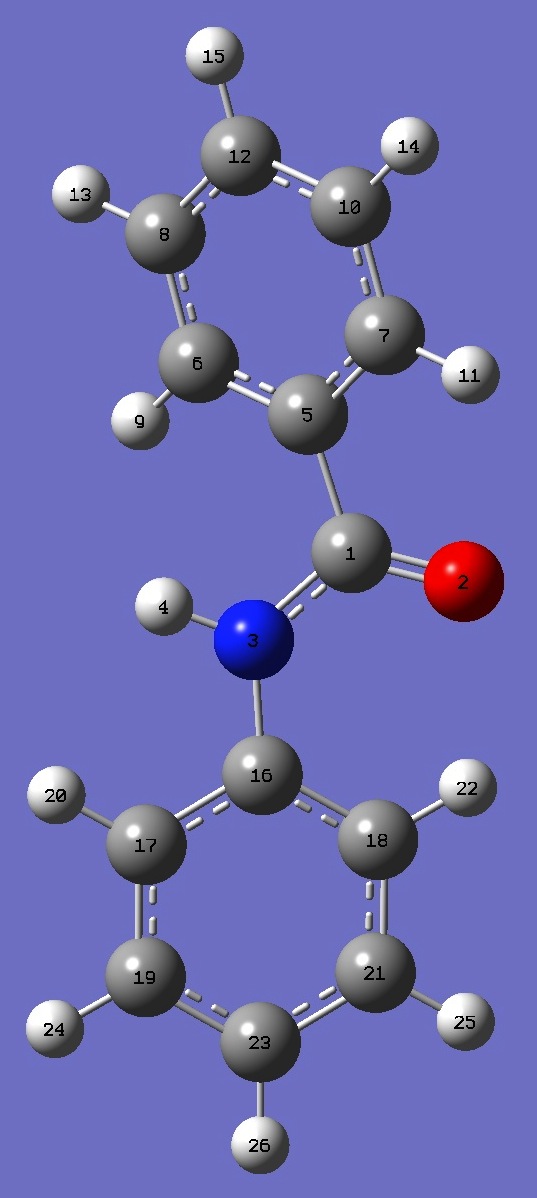
Table 2. Benzanilide.
HF/6-311+G(d,p) and B3LYP/6-311++G(d,p) optimized structure parameters (Å and
degrees).
|
| |
|
|
|
|
|
|
|
|
C
O,1,B1
N,1,B2,2,A1
H,3,B3,1,A2,2,D1,0
C,1,B4,2,A3,3,D2,0
C,5,B5,1,A4,2,D3,0
C,5,B6,1,A5,2,D4,0
C,6,B7,5,A6,1,D5,0
H,6,B8,5,A7,1,D6,0
C,7,B9,5,A8,1,D7,0
H,7,B10,5,A9,1,D8,0
C,10,B11,7,A10,5,D9,0
H,8,B12,6,A11,5,D10,0
H,10,B13,7,A12,5,D11,0
H,12,B14,10,A13,7,D12,0
C,3,B15,1,A14,2,D13,0
C,16,B16,3,A15,1,D14,0
C,16,B17,3,A16,1,D15,0
C,17,B18,16,A17,3,D16,0
H,17,B19,16,A18,3,D17,0
C,18,B20,16,A19,3,D18,0
H,18,B21,16,A20,3,D19,0
C,21,B22,18,A21,16,D20,0
H,19,B23,17,A22,16,D21,0
H,21,B24,18,A23,16,D22,0
H,23,B25,21,A24,18,D23,0
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| HF |
B3LYP |
|
|
|
|
B1=1.19447744
B2=1.36233924
B3=0.991636
B4=1.50490104
B5=1.3893548
B6=1.3896411
B7=1.38569378
B8=1.07539226
B9=1.38289189
B10=1.07345498
B11=1.38668403
B12=1.07507323
B13=1.07507696
B14=1.07538673
B15=1.40961427
B16=1.39272204
B17=1.38964113
B18=1.38121449
B19=1.07702789
B20=1.38675858
B21=1.06869423
B22=1.38320754
B23=1.07548804
B24=1.07562161
B25=1.07478985
A1=124.05409873
A2=116.04168557
A3=120.99029318
A4=123.14930062
A5=117.42507889
A6=120.29350501
A7=120.71884905
A8=120.27315779
A9=118.98125195
A10=120.09042815
A11=119.78454415
A12=119.82205068
A13=120.06869889
A14=128.73280613
A15=116.82498705
A16=123.82322409
A17=120.63053377
A18=119.79122464
A19=119.33111488
A20=120.26909937
A21=121.47598191
A22=119.39456096
A23=118.6609597
A24=120.57431535
D1=170.08547721
D2=179.25969943
D3=149.36915651
D4=-28.72780961
D5=-178.78344192
D6=-0.93724549
D7=179.62511052
D8=-0.26768532
D9=-1.09166668
D10=179.15545842
D11=179.30445556
D12=-179.53228717
D13=-1.40467752
D14=169.51416562
D15=-11.66024993
D16=178.77246548
D17=-1.28285312
D18=-178.74932718
D19=1.69216178
D20=0.00961608
D21=179.98506489
D22=-179.95680224
D23=-179.95554875
|
B1=1.22074603
B2=1.37739865
B3=1.0083648
B4=1.50558675
B5=1.40030555
B6=1.39942513
B7=1.39339266
B8=1.08480735
B9=1.39070945
B10=1.08294093
B11=1.3951552
B12=1.08404888
B13=1.08410252
B14=1.08413861
B15=1.41129932
B16=1.40307646
B17=1.40105259
B18=1.38934156
B19=1.0861604
B20=1.39375583
B21=1.07867232
B22=1.39274784
B23=1.08416987
B24=1.08436901
B25=1.08361817
A1=123.76891736
A2=115.89149114
A3=121.41458575
A4=123.54081092
A5=117.3007147
A6=120.40165919
A7=120.6973854
A8=120.40783912
A9=118.52039061
A10=120.16425403
A11=119.77137967
A12=119.79570731
A13=120.1407567
A14=129.17234081
A15=117.08154368
A16=123.53947949
A17=120.57131015
A18=119.68044185
A19=119.33091596
A20=119.64103172
A21=121.35797004
A22=119.45520422
A23=118.75051201
A24=120.49666649
D1=172.22620831
D2=179.24028862
D3=152.36341529
D4=-25.61087037
D5=-178.84534144
D6=-1.41696005
D7=179.54575068
D8=-0.26241285
D9=-0.94687953
D10=179.15291077
D11=179.34296647
D12=-179.59227649
D13=-2.37418255
D14=175.21139457
D15=-5.35490205
D16=179.35041082
D17=-0.60994069
D18=-179.28104272
D19=1.09243398
D20=-0.08848323
D21=-179.98086738
D22=-179.98906726
D23=-179.93255616
|
|
|
|
|
|
|
|