|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH2=C(H)CH2D
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Deuterium |
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
|
in Propene-3-d1
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Deuterium nqcc's were determined in CH2=C(H)CH2D by Demaison et al. [1], which authors derived also a semi-experimental equilibrium structure (reSE).
|
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the deuterium nqcc's
in S-CH2=C(H)CH2D (D atom in symmetry plane) was made on the reSE equilibrium structure and on an ropt structure given by MP2/aug-cc-pVTZ optimization. Calculated
and experimental deuterium nqcc's are compared in Table 1. Structure parameters
are given
in Table 2. |
|
|
|
|
|
|
|
|
|
|
|
|
In Table 1, subscripts a,b,c refer to the
principal axes of the inertia tensor, subscripts x,y,z to the principal
axes of the nqcc tensor. The nqcc y-axis is chosen coincident
with the inertia c-axis, these are perpendicular to the plane of the
molecule. Ø (degrees) is the angle between its subscripted
parameters. ETA = (Xxx - Xyy)/Xzz. |
|
|
RMS is the root
mean square difference between calculated and experimental diagonal
nqcc's
(percentage of the average of the magnitudes of the experimental
nqcc's), RSD is the calibration residual standard deviation for the B3LYP/6-31G(df,3p) model for
calculation of the efg's/nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
Table 1. D(7) - see figure below for atomic numbering - nqcc's
in S-CH2=C(H)CH2D (kHz). Calculation was made on the rese and ropt structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
Calc /rese |
|
Calc /ropt |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
|
D(7) |
Xaa |
|
- 90.4
|
|
- 90.1
|
|
- 95(4)
|
|
|
|
Xbb |
|
186.9
|
|
186.4 |
|
192(4) |
|
|
|
Xcc |
|
- 96.5
|
|
- 96.3
|
|
- 97(3)
|
|
|
|
|Xab| |
|
- 9.6
|
|
- 9.8
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS |
|
4.0 (3.1 %) |
|
4.3 (3.4 %) |
|
|
|
|
|
RSD |
|
1.1 (0.9 %) |
|
1.1 (0.9 %) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
- 90.7
|
|
- 90.5
|
|
|
|
|
|
Xyy |
|
- 96.5
|
|
- 96.3
|
|
|
|
|
|
Xzz |
|
187.2 |
|
186.8 |
|
|
|
|
|
ETA |
|
0.0308 |
|
0.0309 |
|
|
|
|
|
Øz,b |
|
1.97 |
|
2.03 |
|
|
|
|
|
Øb,CD |
|
1.87 |
|
1.93 |
|
|
|
|
|
Øz,CD |
|
0.10 |
|
0.10
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Table 2. Molecular
structure parameters, rese and ropt = MP2/aug-cc-pVTZ optimization (Å and degrees). |
| |
|
|
|
|
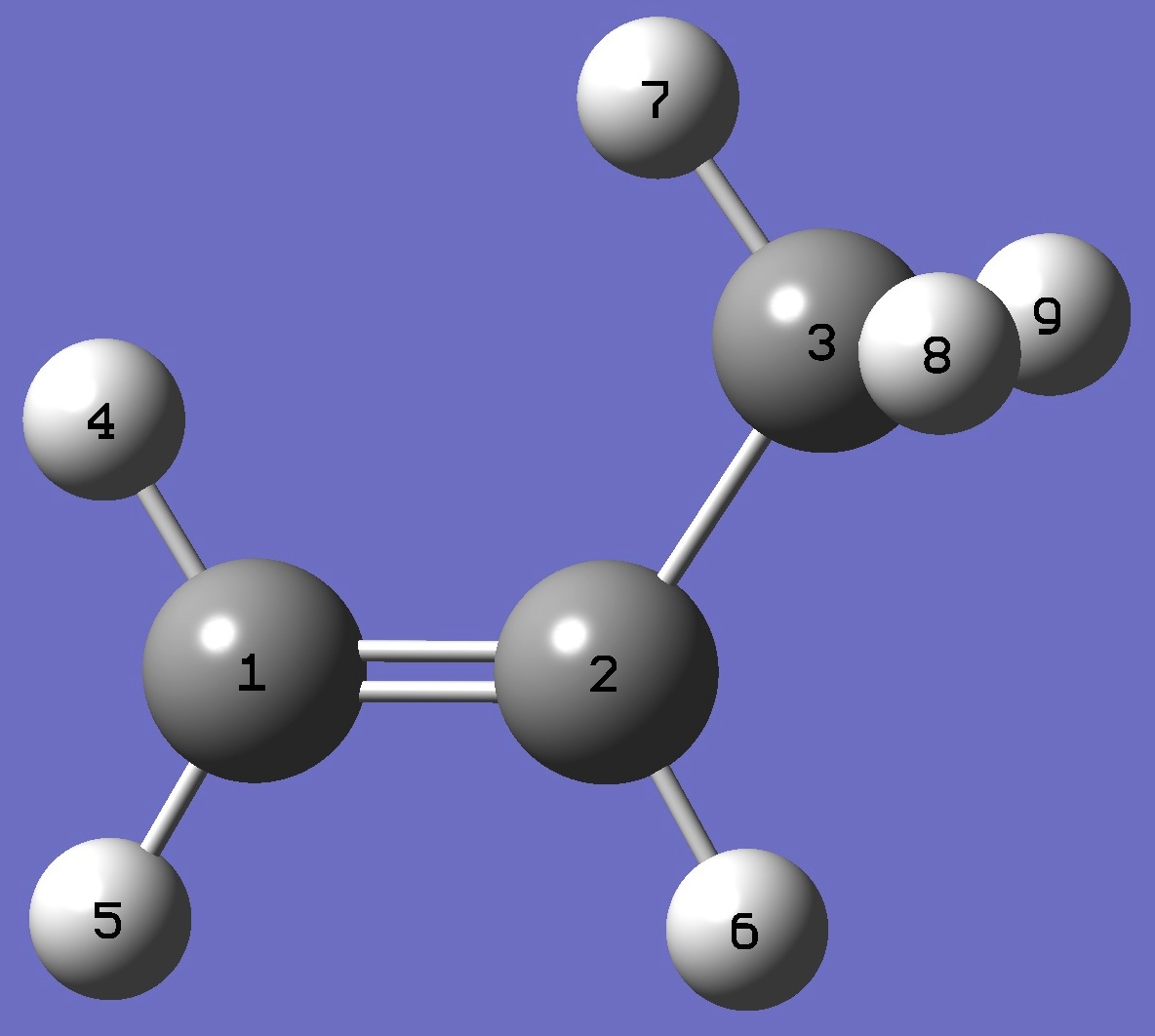 |
C
C 1 R1
C 2 R2 1 A2
H 1 R3 2 A3 3 D3
H 1 R4 2 A4 3 D4
H 2 R5 1 A5 4 D5
H 3 R6 2 A6 1 D6
H 3 R7 2 A7 1 D7
H 3 R8 2 A8 1 D8
|
|
|
|
|
|
|
|
|
rese |
ropt |
|
|
|
|
|
|
|
|
R1=1.33148(26)
R2=1.49530(25)
R3=1.08246(31)
R4=1.08124(26)
R5=1.08497(19)
R6=1.088664(88)
R7=1.09197(16)
R8=1.09197(16)
A2=124.4570(50)
A3=121.154(18)
A4=121.407(32)
A5=118.798(85)
A6=111.025(12)
A7=110.890(21)
A8=110.890(21)
D3=0.
D4=180.
D5=180.
D6=0.
D7=120.627(27)
D8=-120.627(27)
|
R1=1.33495642
R2=1.49523208
R3=1.08273402
R4=1.08070611
R5=1.08514211
R6=1.08910958
R7=1.09092517
R8=1.09092517
A2=124.46566911
A3=121.03457388
A4=121.3592081
A5=118.68505792
A6=111.01233991
A7=110.9197465
A8=110.9197465
D3=0.
D4=180.
D5=180.
D6=0.
D7=120.61324176
D8=-120.61324176
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1]
J.Demaison, N.C.Craig, R.M.Gurusinghe, M.J.Tubergen, H.D.Rudolph,
L.H.Coudert, P.G.Szalay, and A.G.Csázár, J.Phys.Chem. A 121(16),3155(2017).
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3D
|
H2CO |
HOCl |
HC(=O)OH |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Deuterium |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH2CHCH2D.html |
|
|
|
|
|
|
Last
Modified 5 April 2017 |
|
|
|
|
|
|
|
|
|
|

