|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
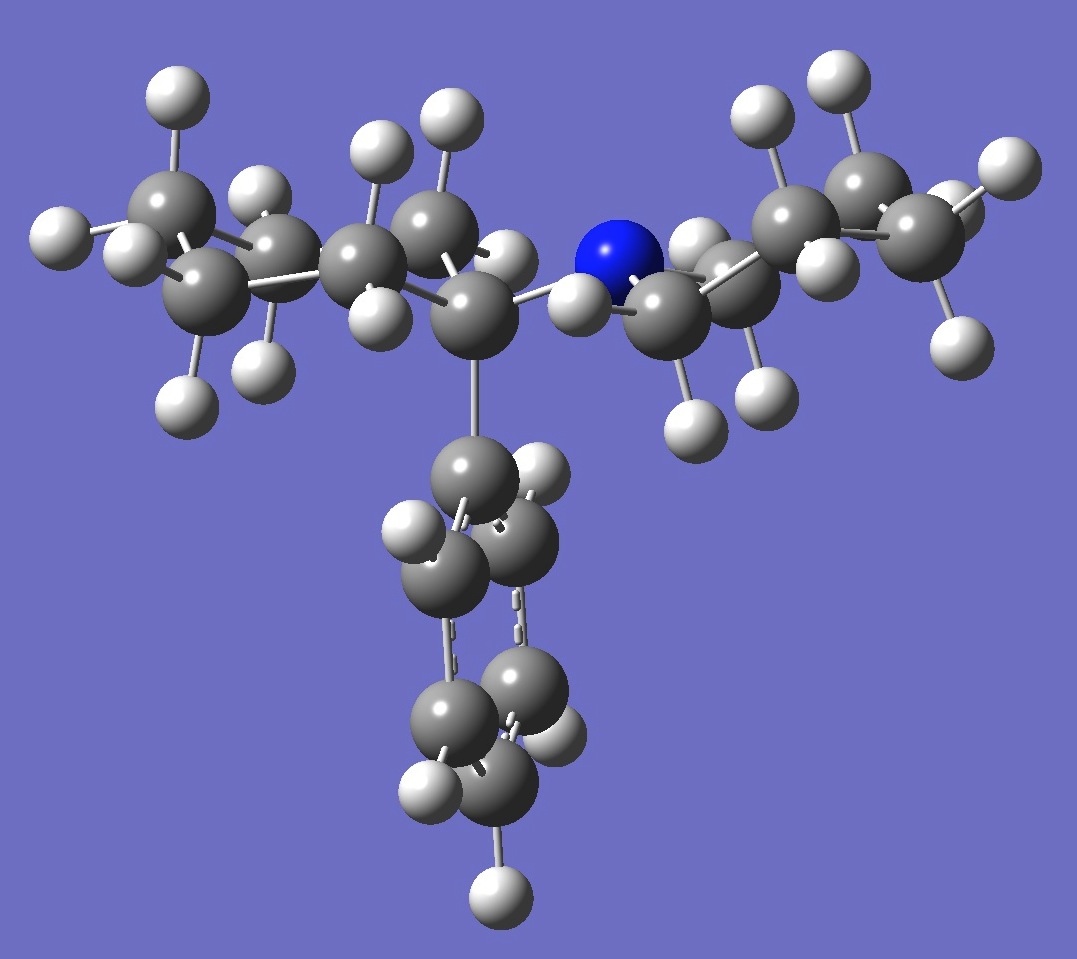
C6H10(-C6H5)NC5H10
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen |
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
in Phencyclidine
|
|
|
|
[1-(1-phenylcyclohexyl)piperidine (PCP) ]
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the 14N
nuclear quadrupole coupling constant tensor in Phencyclidine was made here on an ropt molecular
structure given by B3P86/6-31G(3d,3p)
optimization (assuming Cs symmetry).
|
|
|
This
calculated nqcc tensor is given in Table 1; structure parameters in
Table 2; rotational constants and electric dipole moments in Table 3.
|
|
|
In Table 1, subscripts a,b,c refer to the
principal axes of the inertia tensor; x,y,z to the principal axes
of the nqcc tensor. ETA = (Xxx - Xyy)/Xzz. Ø (degrees) is the angle between its subscripted parameters. RSD is the calibration residual standard
deviation of the B3PW91/6-311+G(df,pd) model for calculation of the efg's/nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
Table 1. Nitrogen nqcc's in Phencyclidine (MHz). Calculation was made
on B3P86/6-31G(3d,3p) ropt structure.
|
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc
|
|
Expt
|
|
| |
|
|
|
|
|
|
|
|
14N |
Xaa |
|
1.984
|
|
|
|
|
|
Xbb |
-
|
4.458
|
|
|
|
|
|
Xcc |
|
2.474
|
|
|
|
|
|
|Xac| |
|
2.652
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RSD
|
0.030 (1.3 %)
|
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
2.935
|
|
|
|
|
|
Xyy |
|
2.474
|
|
|
|
|
|
Xzz |
-
|
5.409
|
|
|
|
|
|
ETA
|
-
|
0.0854
|
|
|
|
|
|
Øz,a
|
|
70.27
|
|
|
|
|
|
Øa,NC(1) * |
|
172.95
|
|
|
|
|
|
Øz,NC(1) *
|
|
102.68
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
* See below for atomic numbering.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
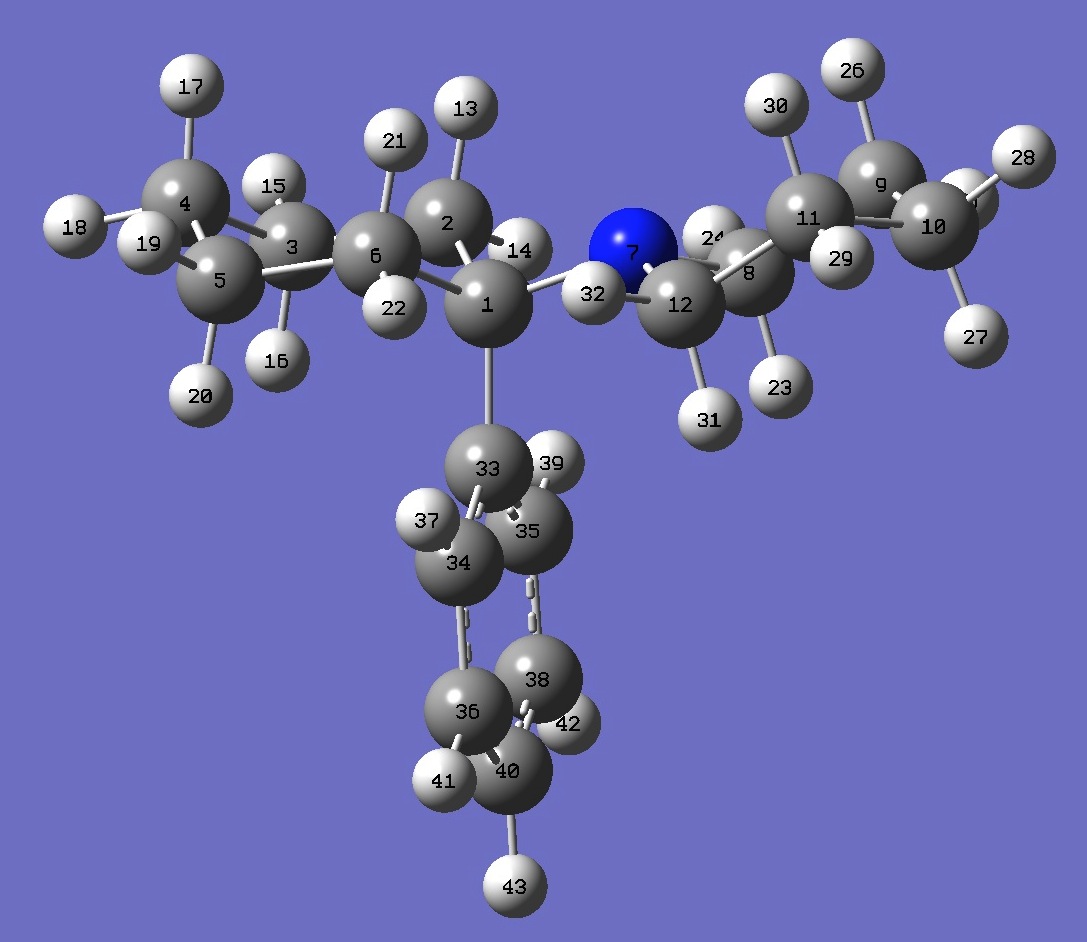
Table 2. Phencyclidine B3P86/6-31G(3d,3p) optimized structure
parameters (Å and degrees).
|
|
|
|
|
|
|
|
|
|
|
|
C
C 1 B1
C 2 B2 1 A1
C 3 B3
2 A2
1 D1 0
C 4 B4
3 A3
2 D2 0
C 5 B5
4 A4
3 D3 0
N 1 B6
6 A5
5 D4 0
C 7 B7
1 A6
6 D5 0
C 8 B8
7 A7
1 D6 0
C 9 B9
8 A8
7 D7 0
C 10 B10
9 A9
8 D8 0
C 7 B11
1 A10 6
D9 0
H 2 B12
1 A11 7
D10 0
H 2 B13
1 A12 7
D11 0
H 3 B14
2 A13 1
D12 0
H 3 B15
2 A14 1
D13 0
H 4 B16
3 A15 2
D14 0
H 4 B17
3 A16 2
D15 0
H 5 B18
4 A17 3
D16 0
H 5 B19
4 A18 3
D17 0
H 6 B20
5 A19 4
D18 0
H 6 B21
5 A20 4
D19 0
H 8 B22
7 A21 1
D20 0
H 8 B23
7 A22 1
D21 0
H 9 B24
8 A23 7
D22 0
H 9 B25
8 A24 7
D23 0
H 10 B26
9 A25 8
D24 0
H 10 B27
9 A26 8
D25 0
H 11 B28 10
A27 9 D26 0
H 11 B29 10
A28 9 D27 0
H 12 B30
7 A29 1
D28 0
H 12 B31
7 A30 1
D29 0
C 1 B32
7 A31 12
D30 0
C 33 B33
1 A32 7
D31 0
C 33 B34
1 A33 7
D32 0
C 34 B35 33
A34 1 D33 0
H 34 B36 33
A35 1 D34 0
C 35 B37 33
A36 1 D35 0
H 35 B38 33
A37 1 D36 0
C 36 B39 34
A38 33 D37 0
H 36 B40 34
A39 33 D38 0
H 38 B41 35
A40 33 D39 0
H 40 B42 36
A41 34 D40 0
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
B1 1.54363931
B2 1.52671953
B3 1.52258398
B4 1.52258398
B5 1.52671953
B6 1.49330547
B7 1.46007899
B8 1.52483578
B9 1.52256072
B10 1.52256072
B11 1.46007899
B12 1.09645449
B13 1.09063897
B14 1.09486167
B15 1.09658714
B16 1.09781249
B17 1.09475902
B18 1.09486167
B19 1.09658714
B20 1.09645449
B21 1.09063897
B22 1.10654125
B23 1.08961760
B24 1.09508230
B25 1.09612524
B26 1.09785032
B27 1.09457024
B28 1.09508230
B29 1.09612524
B30 1.10654125
B31 1.08961760
B32 1.53699400
B33 1.39973015
B34 1.39973015
B35 1.39028476
B36 1.08393777
B37 1.39028476
B38 1.08393777
B39 1.38865518
B40 1.08533698
B41 1.08533698
B42 1.08484750
|
A1 113.96405096
A2 111.58000474
A3 109.99288295
A4 111.58000474
A5 108.06403996
A6 115.45604143
A7 110.92385205
A8 111.07126701
A9 109.48950888
A10 115.45604143
A11 106.33231766
A12 111.73052086
A13 109.07368550
A14 109.62282531
A15 109.38473955
A16 110.66870569
A17 110.57923993
A18 109.41873319
A19 109.83038626
A20 108.67763795
A21 111.45246737
A22 109.61920887
A23 109.08987130
A24 108.69698188
A25 109.29969679
A26 111.00121648
A27 110.88662236
A28 109.75989198
A29 111.45246737
A30 109.61920887
A31 110.73252896
A32 121.46090451
A33 121.46090451
A34 121.67337593
A35 120.05775196
A36 121.67337593
A37 120.05775196
A38 120.35378138
A39 119.50782192
A40 119.50782192
A41 120.49660064
|
D1 57.68474659
D2 -54.03049735
D3 54.03049735
D4 171.15039900
D5 -172.52712227
D6 166.15351425
D7 58.08332679
D8 -52.25519637
D9 58.81257624
D10 -50.01285725
D11 65.17915191
D1 -179.85493010
D13 -63.68725118
D14 66.13082312
D15 -176.62498415
D16 175.62298624
D17 -67.45989386
D18 61.48648027
D19 177.00912571
D20 -72.00934638
D21 46.22736376
D22 -179.39327320
D23 -62.77264176
D24 67.44158341
D25 -175.14902542
D26 173.73266425
D27 -67.97242402
D28 72.00934638
D29 -46.22736376
D30 -64.33015074
D31 87.79052799
D32 -87.79052799
D33 -175.85347149
D34 4.58059440
D35 175.85347149
D36 -4.58059440
D37 0.06666377
D38 -179.79780452
D39 179.79780452
D40 -179.85729748
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table
3. Phencyclidine. Rotational Constants (MHz) and Electric
Dipole Moments (D).
|
|
|
|
|
|
|
|
Calc.
|
Expt
|
|
|
|
|
|
|
|
A
|
515.8
|
|
|
|
B
|
372.0
|
|
|
|
C
|
284.6
|
|
|
|
|
|
|
|
|
|µa|
|
0.11
|
|
|
|
|µb| |
0.13
|
|
|
|
|µc| |
0 (symmetry)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Piperidine |
N-Methylpiperidine | N-Cyclohexylpiperidine |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
PCP.html |
|
|
|
|
|
|
Last
Modified 25 Oct 2013 |
|
|
|
|
|
|
|
|
|
|

