|
|
|
|
|
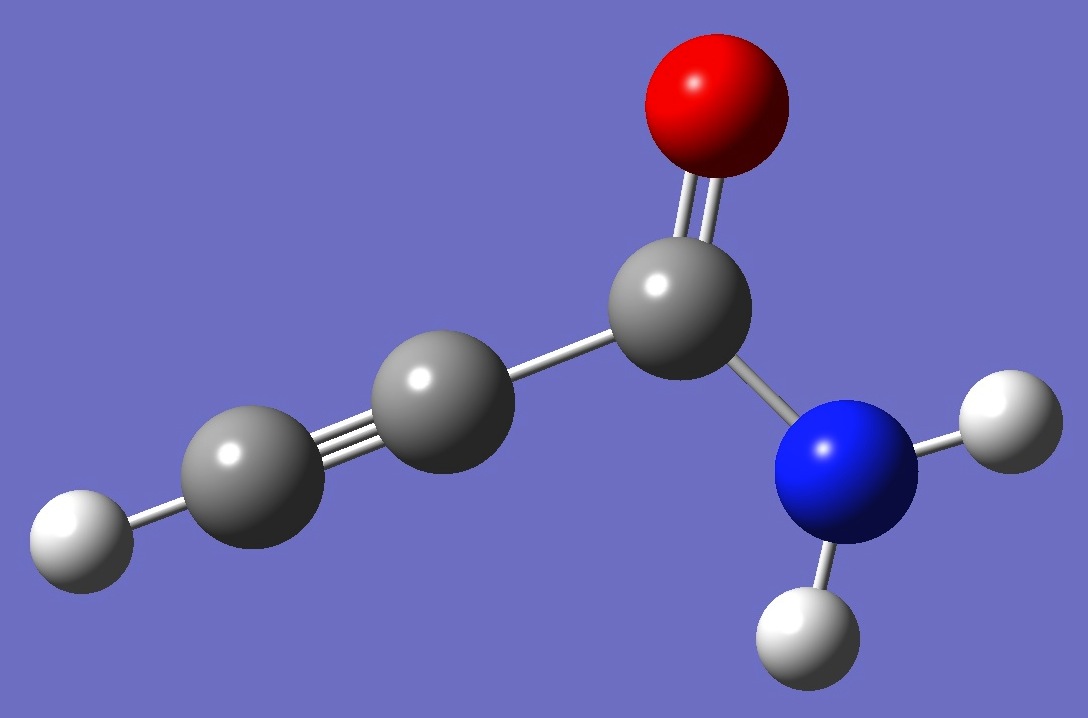
| Table 2. Propiolamide. Molecular structure parameters (Å
and degrees). |
| |
|
|
|
|
ropt (1) = HF/6-311++G(3df,3pd) optimization. |
|
ropt (2) = HF/aug-cc-pVTZ(G03) optimization. |
| |
|
|
|
| Point Group: Cs |
|
|
|
|
|
|
|
|
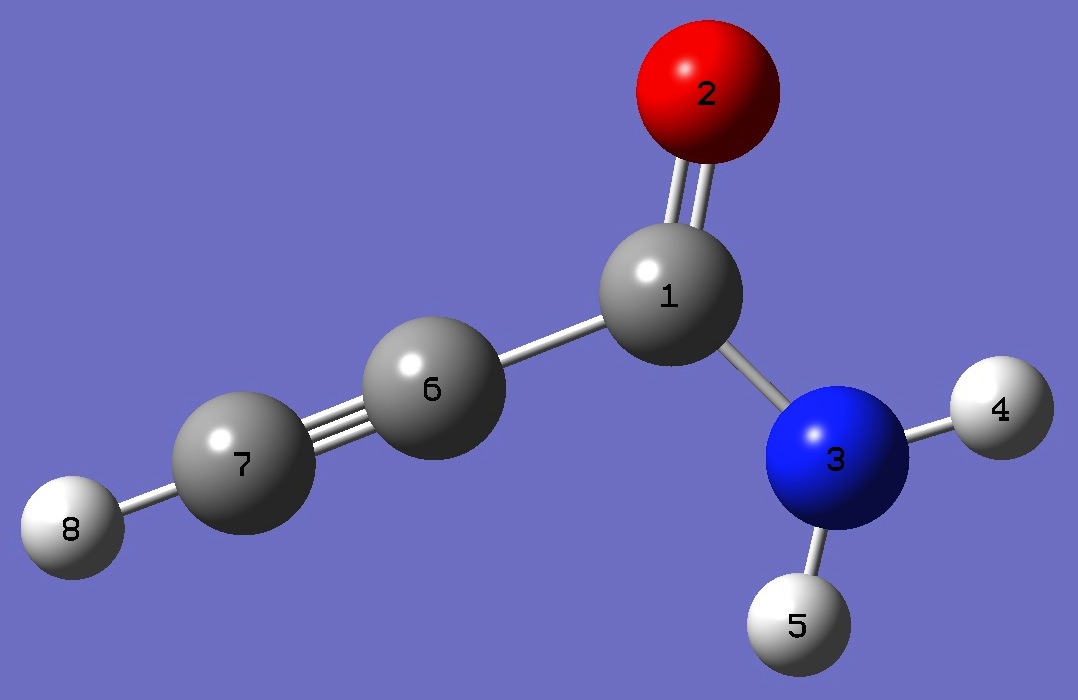
C
O,1,B1
N,1,B2,2,A1
H,3,B3,1,A2,2,D1,0
H,3,B4,1,A3,2,D2,0
C,1,B5,2,A4,3,D3,0
C,6,B6,2,A5,1,D4,0
H,7,B7,3,A6,1,D5,0
|
|
|
|
|
|
|
|
|
|
| ropt (1) |
ropt (2) |
|
|
|
|
|
|
B1=1.1866456
B2=1.3459237
B3=0.99123019
B4=0.98889444
B5=1.46927606
B6=1.17986
B7=1.05448465
A1=124.17445886
A2=118.43762231
A3=121.62714349
A4=122.00173871
A5=154.41298543
A6=159.76537472
D1=0.
D2=180.
D3=180.
D4=180.
D5=180.
|
B1=1.18825324
B2=1.34630552
B3=0.99104866
B4=0.98861777
B5=1.46903403
B6=1.18008682
B7=1.05446413
A1=124.17141716
A2=118.46896821
A3=121.59883644
A4=122.03592832
A5=154.32250622
A6=159.77220779
D1=0.
D2=180.
D3=180.
D4=180.
D5=180.
|
|
|
|
|
|
|
|