|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
CH3CH2-C(=O)-NH2
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Nitrogen |
|
|
|
Nuclear
Quadrupole Coupling Constants |
|
|
in Propionamide
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Calculation of the
nitrogen nqcc's in propionamide was made here on molecular
structures given by HF/6-311++G(3df,3pd) and HF/aug-cc-pVTZ(G03) optimizations, with Cs symmetry assumed. These are
compared with the
experimental nqcc's [1] in Table 1. Structure parameters are
given in Table 2, rotational constants in Table 3.
|
|
|
|
|
|
|
|
|
|
|
|
|
In Table 1, subscripts a,b,c refer to the
principal axes of the inertia tensor; x,y,z to the principal axes
of the nqcc tensor.
ě (degrees) is the angle between its subscripted
parameters. ETA = (Xxx - Xyy)/Xzz. |
|
|
|
|
|
|
|
|
|
|
|
|
RSD is the
calibration residual standard deviation of the B3PW91/6-311+G(df,pd) model for calculation of nitrogen efg's/nqcc's. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
Table 1. 14N nqcc's in Propionamide (MHz). Calculation was made
on the (1) HF/6-311++G(3df,3pd) and (2) HF/aug-cc-pVTZ(G03) optimized structures. |
|
| |
|
|
|
|
|
|
|
|
|
|
|
Calc. (1)
|
|
Calc. (2) |
|
Expt. [1] |
|
| |
|
|
|
|
|
|
|
|
|
Xaa |
|
2.071 |
|
2.071 |
|
2.0341(33) |
|
|
Xbb |
|
2.073 |
|
2.070 |
|
1.9421(35) |
|
|
Xcc |
- |
4.144 |
- |
4.141 |
-
|
3.9762
|
|
|
|Xab| |
|
0.035 |
|
0.038 |
|
|
|
|
|
|
|
|
|
|
|
|
|
RMS
|
|
0.125 (4.7 %)
|
|
0.122 (4.6 %)
|
|
|
|
|
RSD |
|
0.030 (1.3 %) |
0.030 (1.3 %) |
|
|
|
|
|
|
|
|
|
|
|
|
Xxx |
|
2.037 |
|
2.033 |
|
|
|
|
Xyy |
|
2.107 |
|
2.108 |
|
|
|
|
Xzz |
- |
4.144 |
- |
4.141 |
|
|
|
|
ETA |
|
0.017 |
|
0.018 |
|
|
|
|
ěy,a |
|
45.72 |
|
44.74 |
|
|
|
|
ěa,CN |
|
30.80 |
|
30.84 |
|
|
|
|
ěy,CN |
|
14.92 |
|
13.90 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table 2. Propionamide. Molecular structure parameters (┼
and degrees). Complete structure is given here in Z-matrix format.
|
| |
|
|
|
|
ropt (1) = HF/6-311++G(3df,3pd) optimization. |
|
ropt (2) = HF/aug-cc-pVTZ(G03) optimization. |
| |
|
|
|
| Point Group: Cs |
|
ropt (1) |
ropt (2) |
|
|
|
|
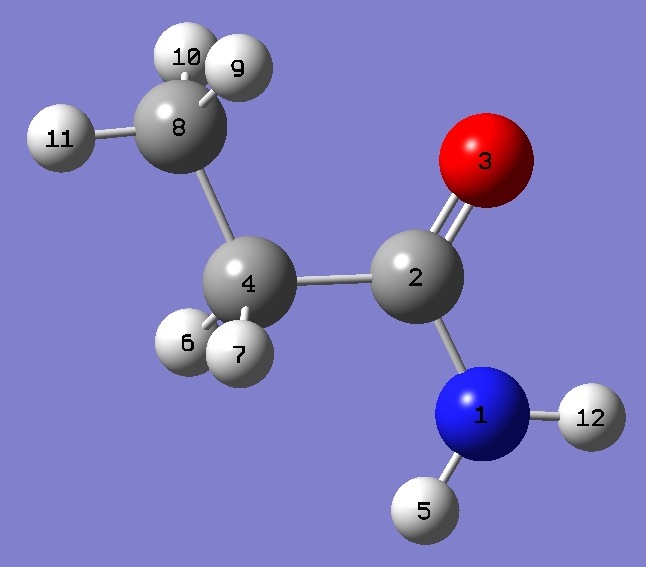
|
NC(2) |
1.3544 |
1.3547 |
| C(2)O |
1.1917 |
1.1933 |
| C(2)C(4) |
1.5173 |
1.5172 |
| C(4)C(8) |
1.5204 |
1.5204 |
| NH(5) |
0.9884 |
0.9882 |
| NH(12) |
0.9909 |
0.9907 |
| C(2)NH(5) |
122.19 |
122.18 |
| C(2)NH(12) |
118.70 |
118.70 |
|
NCO |
121.89 |
121.87 |
| Ethyl hydrogens? |
NC(2)C(4) |
114.83 |
114.84 |
| See Z-Matrix |
C(2)C(4)C(8) |
113.26 |
113.32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
| Table 3. Propionamide. Rotational Constants (MHz). Normal Species. |
| |
|
|
|
|
|
ropt (1) = HF/6-311++G(3df,3pd) optimization. |
|
ropt (2) = HF/aug-cc-pVTZ(G03) optimization. |
| |
|
|
|
|
|
|
Calc. ropt (1) |
Calc. ropt (2) |
Expt. [1] |
|
|
|
|
|
|
A |
10206.1 |
10192.2 |
9528.7(10)
|
|
B |
3813.4 |
3810.8 |
4139.9(10)
|
|
C |
2872.6 |
2870.0 |
2850.3865(53)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
[1]
A.A.Mescheryakov, E.A.Alekseev, V.V.Ilyushin, R.A.Motiyenko,
L.MargulÚs, F.J.Lovas, Abstract RJ04, 69th International Symposium on
Molecular Spectroscopy, Champaign- Urbana, Ill. 2014.
|
|
|
|
|
|
|
|
|
|
|
|
|
K.-M.Marstokk, H.M°llendal, and S.Samdal, J.Mol.Struct. 376,11(1996): Xaa = 2.2(8), Xbb = 2.3(5) MHz.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Formamide |
Acetamide |
N-Ethylformamide |
|
|
|
N-Acetylglycine |
Propiolamide |
|
|
|
|
N-Methylacetamide |
N-Methylpropionamide |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Table of Contents |
|
|
|
|
|
Molecules/Nitrogen |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Propionamide.html |
|
|
|
|
|
|
Last
Modified 24 May 2015 |
|
|
|
|
|
|
|
|
|
|

