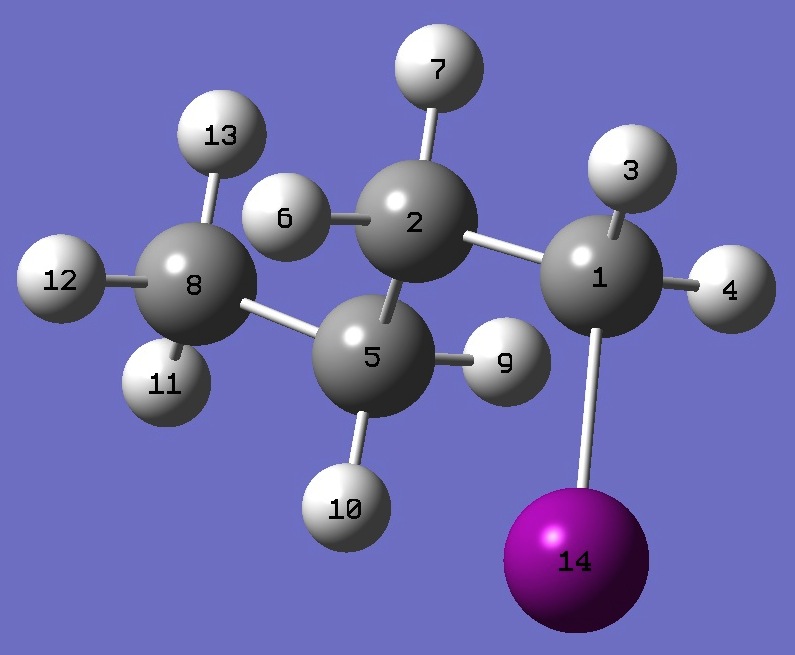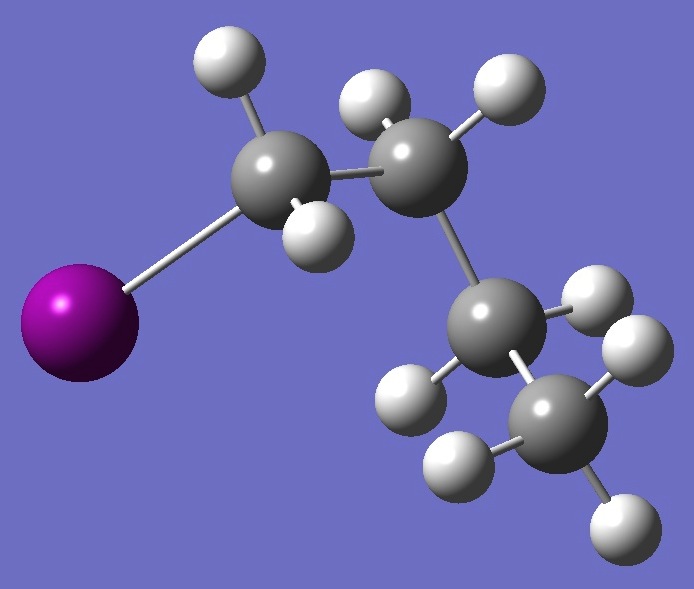
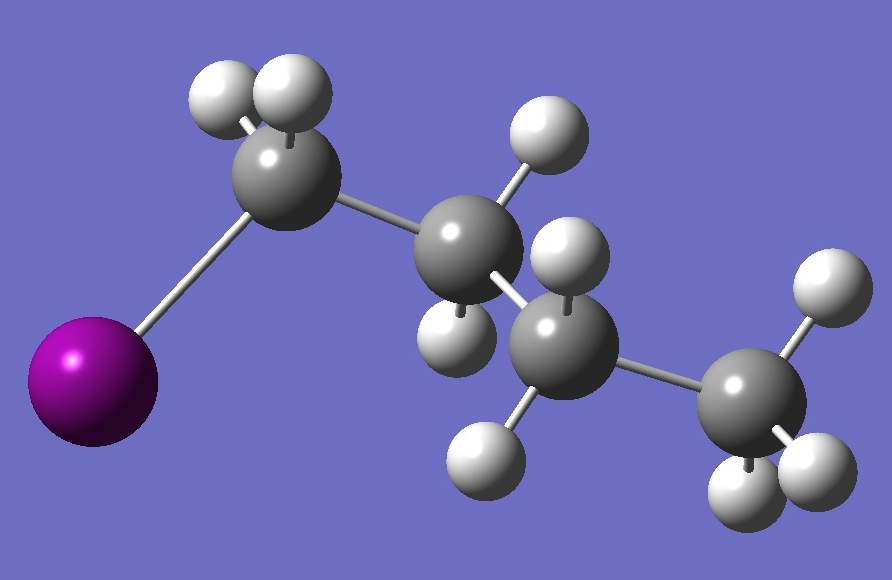
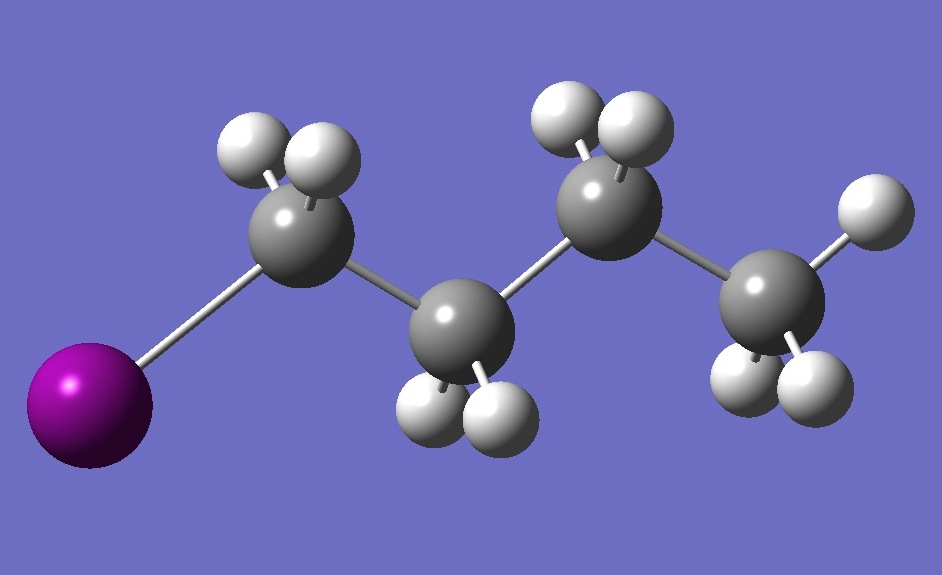
| |
||||||||
| Table 1. 127I nqcc's in GA-1-Iodobutane (MHz). Calculation was made on the following structures: ropt (1) = MP2/6-311+G(d,p) optimization, and ropt (2) = MP2/6-311+G(3df,3pd) optimization. | ||||||||
| |
||||||||
| Calc. (1) |
Calc. (2) | Expt. [1] | ||||||
| |
||||||||
| Xaa | - 545.9 |
- 496.6 |
- 521.973(12) |
|||||
| Xbb | - 314.1 |
- 350.9 |
- 337.242(15) |
|||||
| Xcc | 860.0 |
847.5 |
859.216(19) |
|||||
| Xab | - |
1335.8 |
- |
1322.6 |
- |
1331.251(23) |
||
| Xac | - 233.8 |
- 225.6 |
- 226.66(13) |
|||||
| Xbc | - 238.4 |
- 239.2 |
- 235.95(6) |
|||||
| RMS | 19.2 (3.36 %) |
17.9 (3.13 %) |
||||||
| RSD | 15.2 (1.23 %) | 15.2 (1.23 %) | ||||||
| Xxx | 888.0 |
875.1 |
884.09(11) |
|||||
| Xyy | 924.4 |
914.1 |
920.04(9) |
|||||
| Xzz | - |
1812.4 |
- |
1789.2 |
- |
1804.133(35) |
||
| ETA | 0.0201 |
0.0218 |
0.01993(8) |
|||||
| Øz,CI | 0.75 |
0.80 |
||||||
| Table 3. GA-1-Iodobutane. Rotational Constants (MHz). ropt (1) = MP2/6-311+G(d,p) and ropt (2) = MP2/6-311+G(3df,3pd) optimization. | ||||
| ropt(1) | ropt(2) | Expt. [1] | ||
| A | 7629. |
7590. |
7532.3121(18) |
|
| B | 960. |
988. |
970.51215(26) |
|
| C | 888. |
912. |
896.60780(18) |
|
| Table 4. GA-1-Iodobutane. Quartic Centrifugal Distortion Constants (kHz). Calc = B3LYP/6-311+G(d,p). |
||||||
| Calc | Expt [1] | |||||
| D_J |
0.200 |
0.2058(24) |
||||
| D_JK |
- |
4.43 |
- |
4.450(18) |
||
| D_K |
50.0 |
45.8(4) |
||||
| d_1 |
- |
0.0305 |
- |
0.0302(6) |
||
| d_2 |
- |
0.000559 |
0.0122(14) |
|||