|
| |
|
|
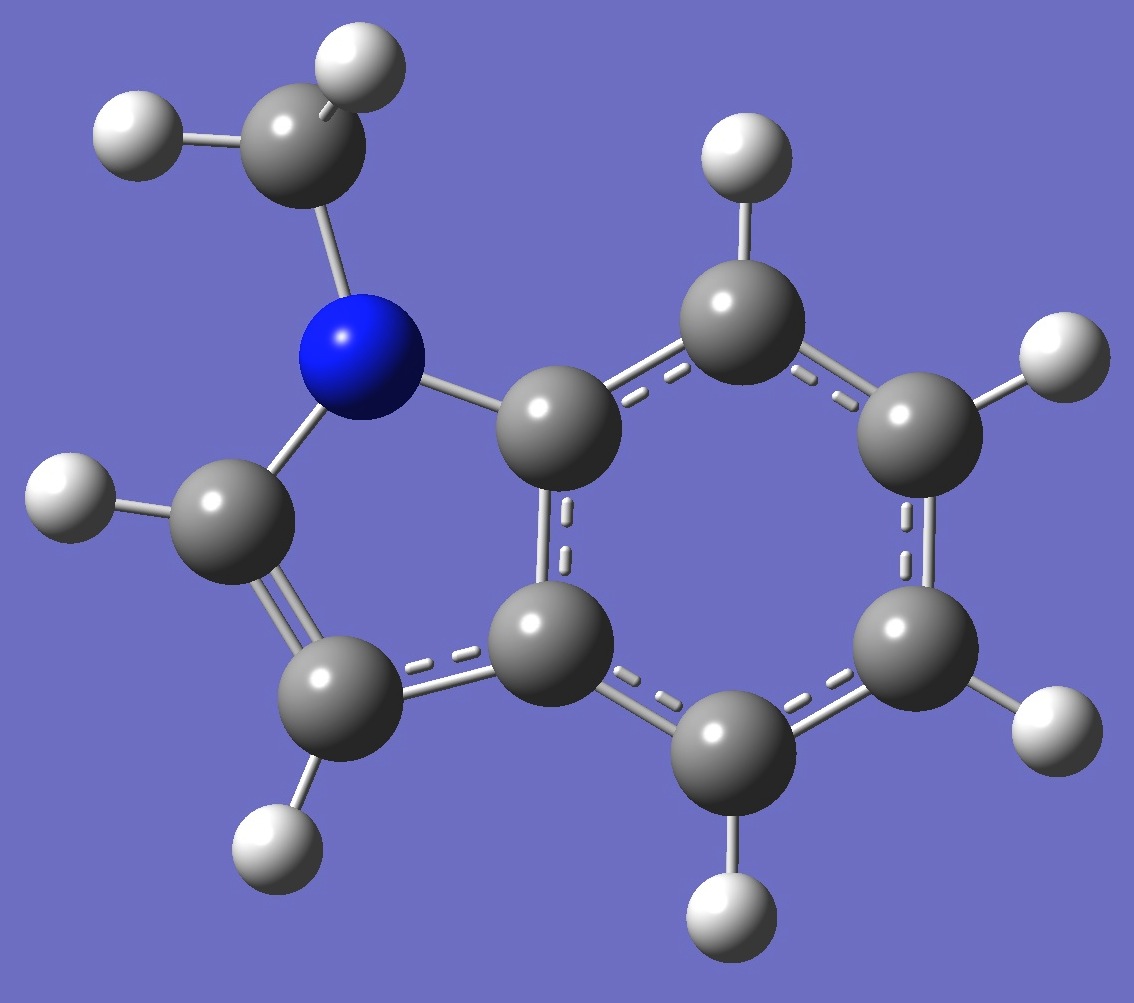
| Table 2. 1-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
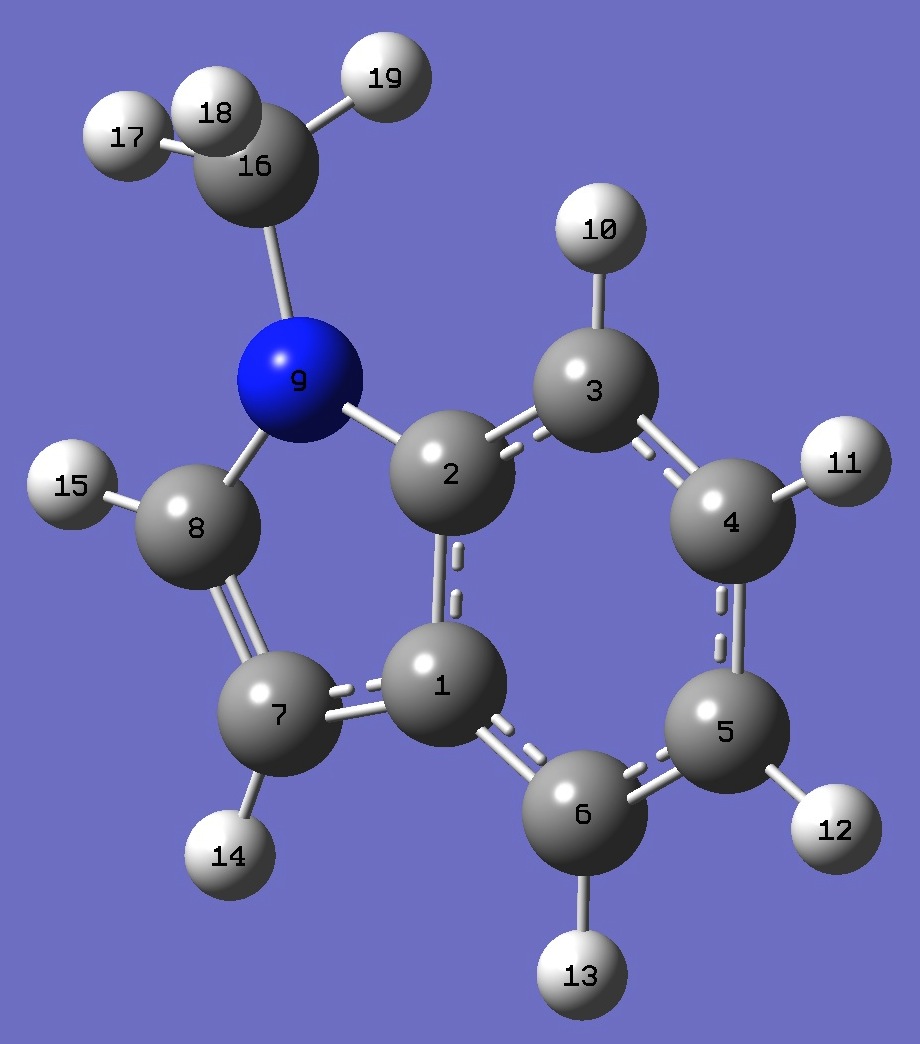
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,4,B10,3,A9,2,D8,0
H,5,B11,4,A10,3,D9,0
H,6,B12,5,A11,4,D10,0
H,7,B13,1,A12,6,D11,0
H,8,B14,7,A13,1,D12,0
C,9,B15,2,A14,1,D13,0
H,16,B16,9,A15,2,D14,0
H,16,B17,9,A16,2,D15,0
H,16,B18,9,A17,2,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41966702
B2=1.39484673
B3=1.38484316
B4=1.40465132
B5=1.38330361
B6=1.43126707
B7=1.36520708
B8=1.37763106
B9=1.08165133
B10=1.08170256
B11=1.08177068
B12=1.08232983
B13=1.07638068
B14=1.07716044
B15=1.44446122
B16=1.08733428
B17=1.09196722
B18=1.09196722
A1=122.13526131
A2=117.56568204
A3=121.21161876
A4=121.12722443
A5=134.46416519
A6=106.8658327
A7=107.79102933
A8=121.58722152
A9=119.40815174
A10=119.18156485
A11=120.48374951
A12=127.23287441
A13=129.82780313
A14=125.55882545
A15=109.09321807
A16=111.13820156
A17=111.13820156
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=0.
D12=180.
D13=180.
D14=180.
D15=-60.47120422
D16=60.47120422
|
B1=1.41878682
B2=1.39418467
B3=1.38480815
B4=1.40446945
B5=1.38361733
B6=1.42993286
B7=1.36628458
B8=1.3745541
B9=1.08503824
B10=1.08481875
B11=1.08485342
B12=1.08528621
B13=1.07945761
B14=1.08059936
B15=1.43926755
B16=1.08992777
B17=1.09435133
B18=1.09435133
A1=122.37476282
A2=117.3927095
A3=121.24316847
A4=121.19537888
A5=134.54730317
A6=106.76383883
A7=107.80745418
A8=121.58812178
A9=119.38213887
A10=119.15533876
A11=120.52454146
A12=127.31270399
A13=130.07331513
A14=125.36366889
A15=109.03097701
A16=111.10002617
A17=111.10002617
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=0.
D12=180.
D13=180.
D14=180.
D15=-60.49876387
D16=60.49876387
|
|
|
|
|