|
| |
|
|
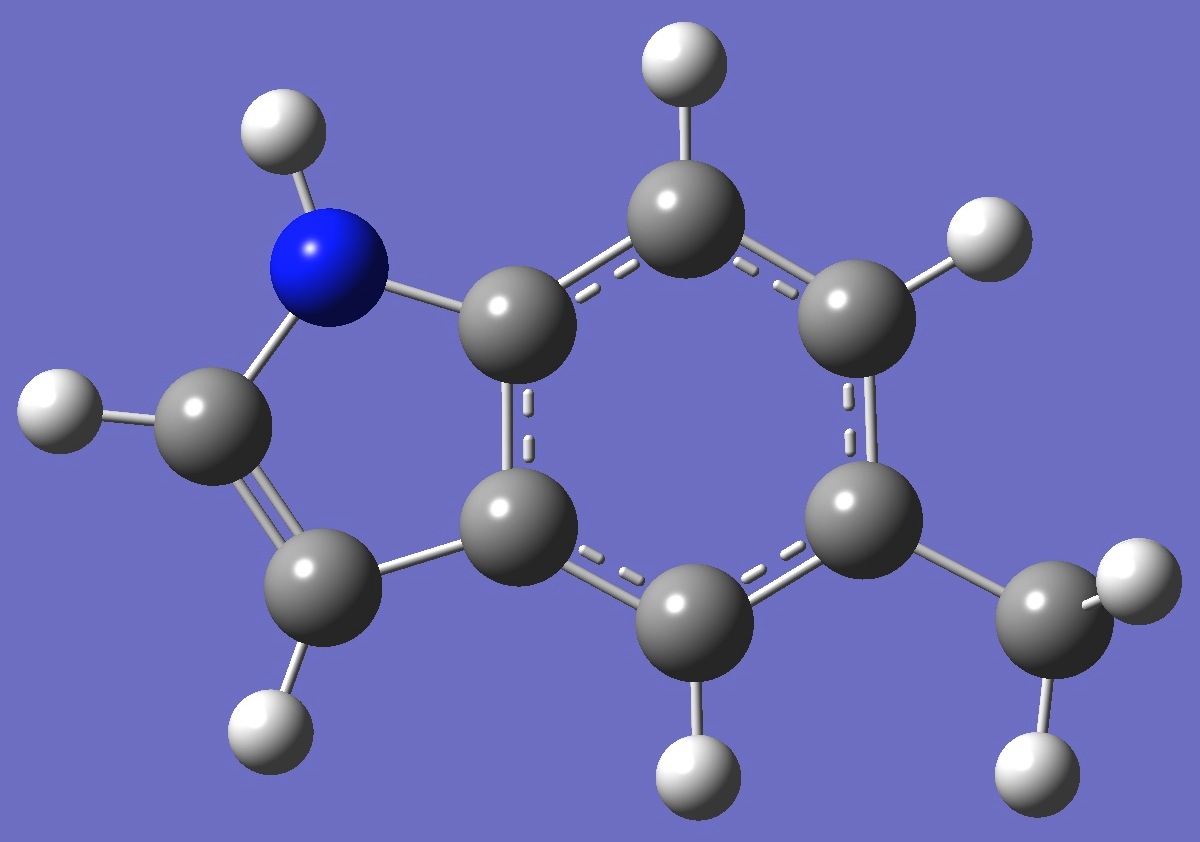
| Table 2. 5-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|

|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,4,B10,3,A9,2,D8,0
H,6,B11,5,A10,4,D9,0
H,7,B12,1,A11,6,D10,0
H,8,B13,7,A12,1,D11,0
H,9,B14,2,A13,1,D12,0
C,5,B15,4,A14,3,D13,0
H,16,B16,5,A15,4,D14,0
H,16,B17,5,A16,4,D15,0
H,16,B18,5,A17,4,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.4155961
B2=1.39343069
B3=1.38235341
B4=1.41178653
B5=1.38507822
B6=1.43293665
B7=1.36561975
B8=1.37743352
B9=1.08250692
B10=1.08302626
B11=1.08330541
B12=1.07650831
B13=1.07657521
B14=1.00293934
B15=1.50817296
B16=1.09264491
B17=1.09264491
B18=1.08964829
A1=121.77341424
A2=117.66984693
A3=122.20099961
A4=119.32242025
A5=134.27456377
A6=107.13066883
A7=107.29402091
A8=121.49385416
A9=118.98414996
A10=119.89109266
A11=127.07965654
A12=129.92757801
A13=125.56206607
A14=119.57247828
A15=111.42610065
A16=111.42610065
A17=111.29590344
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=0.
D11=180.
D12=180.
D13=180.
D14=-59.73240193
D15=59.73240193
D16=180.
|
B1=1.41546289
B2=1.39342923
B3=1.3822325
B4=1.41138462
B5=1.38549834
B6=1.43130572
B7=1.36664373
B8=1.37489599
B9=1.08535344
B10=1.08614596
B11=1.08635252
B12=1.07954902
B13=1.07979078
B14=1.00479355
B15=1.5037349
B16=1.09493183
B17=1.09493183
B18=1.09209154
A1=121.96786976
A2=117.51099031
A3=122.21802675
A4=119.43040123
A5=134.32213189
A6=107.02344258
A7=107.28854941
A8=121.56053945
A9=119.05204104
A10=119.85972936
A11=127.18547279
A12=130.0731547
A13=125.45129001
A14=119.55353665
A15=111.45987846
A16=111.45987846
A17=111.24780513
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=0.
D11=180.
D12=180.
D13=180.
D14=-59.74485499
D15=59.74485499
D16=180.
|
|
|
|
|