|
| |
|
|
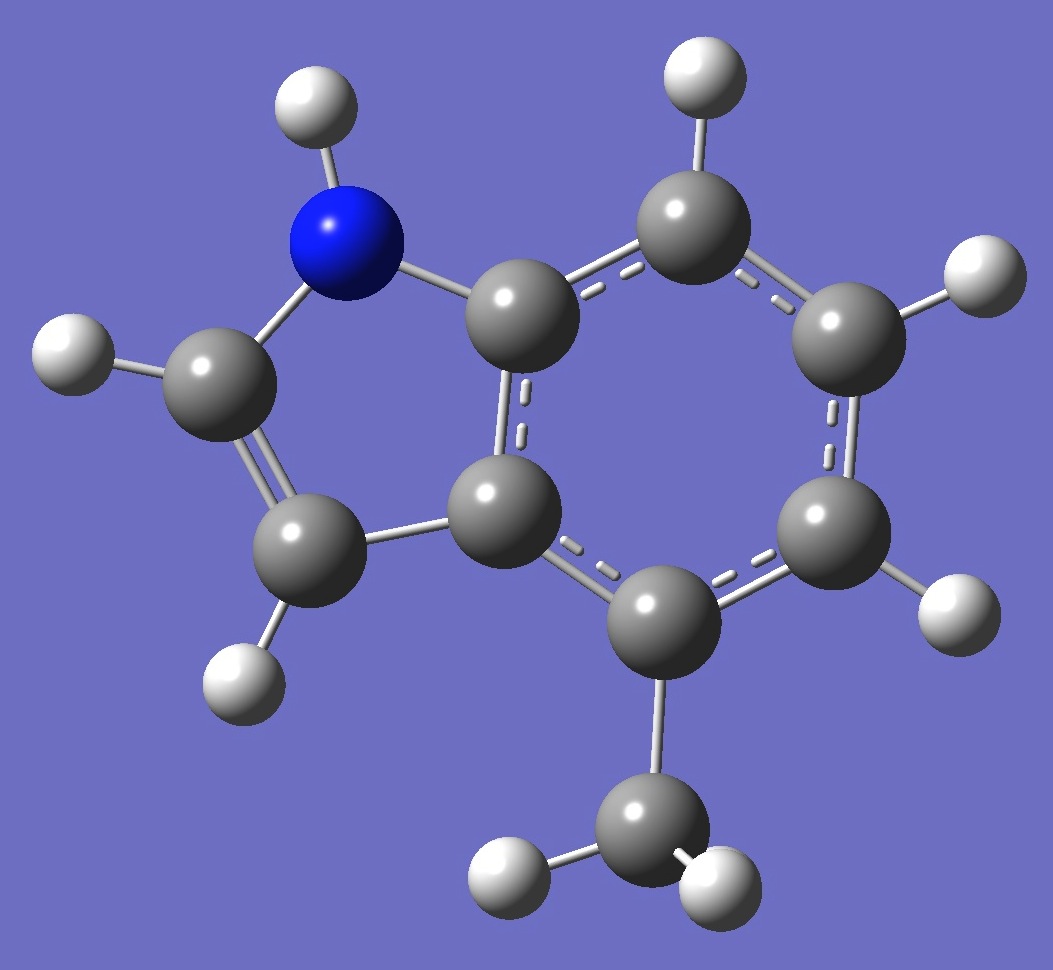
| Table 2. 4-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
|
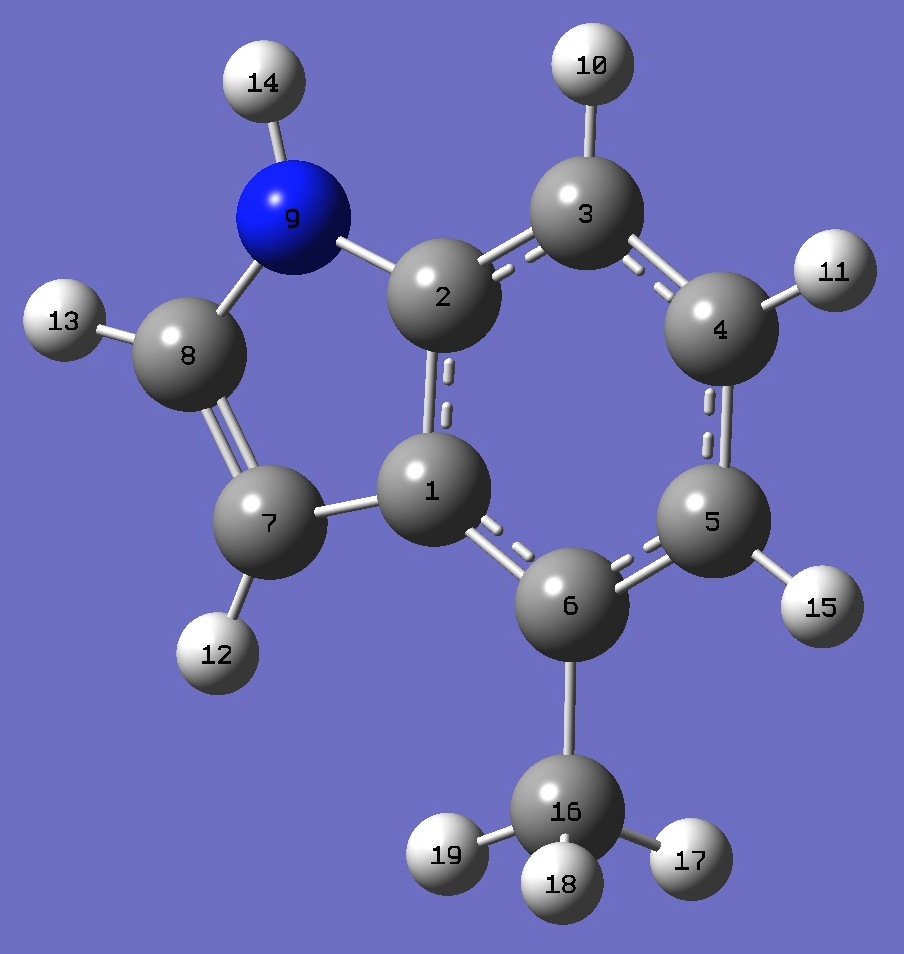
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,4,B10,3,A9,2,D8,0
H,7,B11,1,A10,6,D9,0
H,8,B12,7,A11,1,D10,0
H,9,B13,2,A12,1,D11,0
H,5,B14,4,A13,3,D12,0
C,6,B15,5,A14,4,D13,0
H,16,B16,6,A15,5,D14,0
H,16,B17,6,A16,5,D15,0
H,16,B18,6,A17,5,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41973622
B2=1.39257468
B3=1.38361257
B4=1.40272614
B5=1.38798782
B6=1.4361605
B7=1.36388915
B8=1.37609272
B9=1.08209916
B10=1.08183128
B11=1.07592154
B12=1.07651152
B13=1.00298665
B14=1.08334615
B15=1.50699398
B16=1.09212147
B17=1.09212147
B18=1.08829323
A1=122.52987665
A2=117.01517358
A3=121.27675066
A4=122.26254003
A5=133.86469443
A6=107.25486028
A7=107.25245507
A8=121.65315671
A9=119.51080735
A10=127.47144846
A11=129.92142155
A12=125.37699279
A13=118.71109626
A14=120.92122536
A15=111.12258596
A16=111.12258596
A17=111.92392176
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=0.
D10=180.
D11=180.
D12=180.
D13=180.
D14=-59.61336714
D15=59.61336714
D16=180.
|
B1=1.41892659
B2=1.39248842
B3=1.38385929
B4=1.40248887
B5=1.38823659
B6=1.43385094
B7=1.36509965
B8=1.37388271
B9=1.0849851
B10=1.08491856
B11=1.07927348
B12=1.07972128
B13=1.0048088
B14=1.08639434
B15=1.50245437
B16=1.09433713
B17=1.09433713
B18=1.09107479
A1=122.63132815
A2=116.9124712
A3=121.31837739
A4=122.25863797
A5=133.79125666
A6=107.12141164
A7=107.22179314
A8=121.70992011
A9=119.47330341
A10=127.5686892
A11=130.07494229
A12=125.28495066
A13=118.81304843
A14=121.03268059
A15=111.19221461
A16=111.19221461
A17=111.78218202
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=0.
D10=180.
D11=180.
D12=180.
D13=180.
D14=-59.65588572
D15=59.65588572
D16=180.
|
|
|
|
|