|
| |
|
|
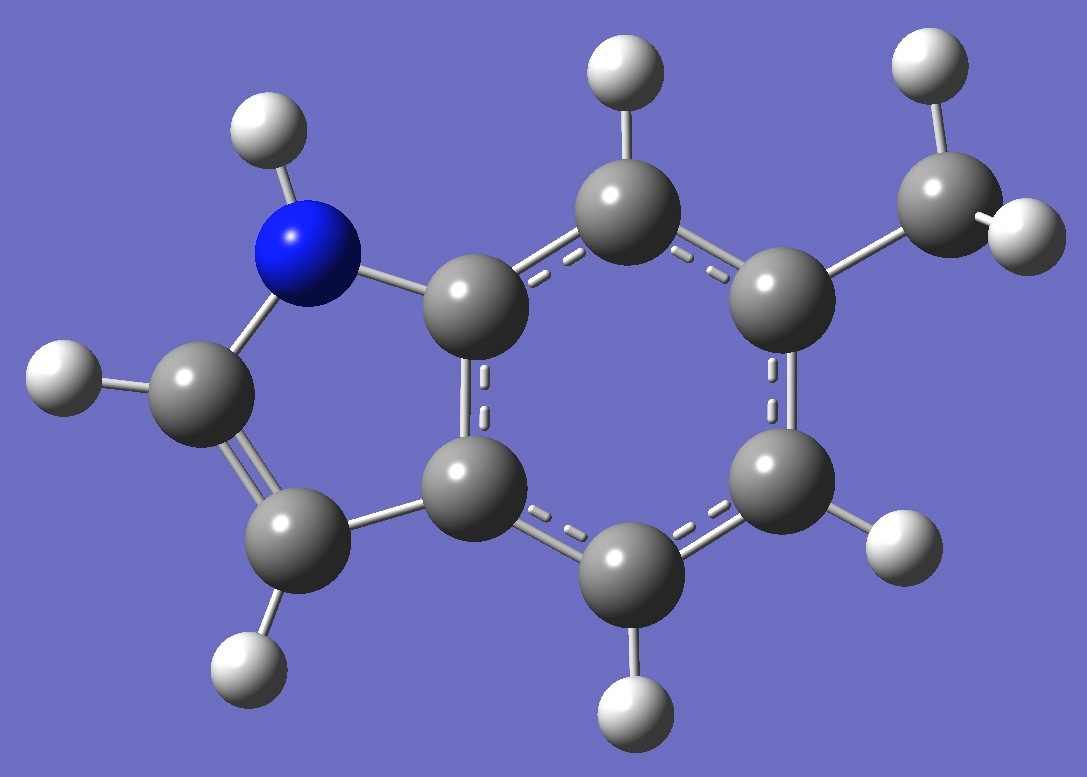
| Table 2. 6-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
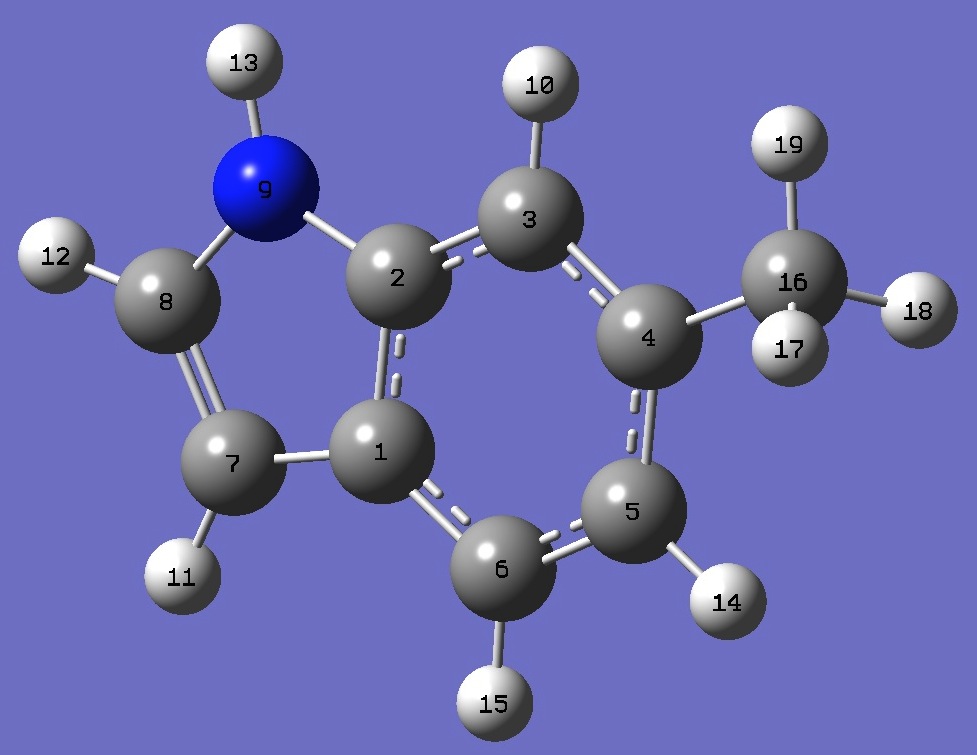
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,7,B10,1,A9,6,D8,0
H,8,B11,7,A10,1,D9,0
H,9,B12,2,A11,1,D10,0
H,5,B13,4,A12,3,D11,0
H,6,B14,5,A13,4,D12,0
C,4,B15,3,A14,2,D13,0
H,16,B16,4,A15,3,D14,0
H,16,B17,4,A16,3,D15,0
H,16,B18,4,A17,3,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41571841
B2=1.39498879
B3=1.38639301
B4=1.41196984
B5=1.38074216
B6=1.43354008
B7=1.36453135
B8=1.37604284
B9=1.0833291
B10=1.07651286
B11=1.07650807
B12=1.00298148
B13=1.08306631
B14=1.0826113
B15=1.50732812
B16=1.09254157
B17=1.09254157
B18=1.08978964
A1=122.37419015
A2=118.59253541
A3=119.37808175
A4=122.12303465
A5=134.76940622
A6=107.08714788
A7=107.15937932
A8=120.9740245
A9=127.08754299
A10=130.01985029
A11=125.48976112
A12=118.59247906
A13=120.28569303
A14=120.86338139
A15=111.31736067
A16=111.31736067
A17=111.47913102
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=0.
D9=180.
D10=180.
D11=180.
D12=180.
D13=180.
D14=120.35749163
D15=-120.35749163
D16=0.
|
B1=1.41556702
B2=1.39445701
B3=1.38662449
B4=1.41155044
B5=1.38090409
B6=1.43179958
B7=1.36568699
B8=1.37376142
B9=1.08630797
B10=1.07956216
B11=1.07973711
B12=1.00483383
B13=1.08620952
B14=1.08546779
B15=1.5030844
B16=1.09483502
B17=1.09483502
B18=1.0922528
A1=122.49943097
A2=118.42257627
A3=119.491168
A4=122.140748
A5=134.74803569
A6=106.98277309
A7=107.16255083
A8=121.13813803
A9=127.19422427
A10=130.15777459
A11=125.37397803
A12=118.5007841
A13=120.34251868
A14=120.76119617
A15=111.35775223
A16=111.35775223
A17=111.43620463
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=0.
D9=180.
D10=180.
D11=180.
D12=180.
D13=180.
D14=120.34082851
D15=-120.34082851
D16=0.
|
|
|
|
|