|
| |
|
|
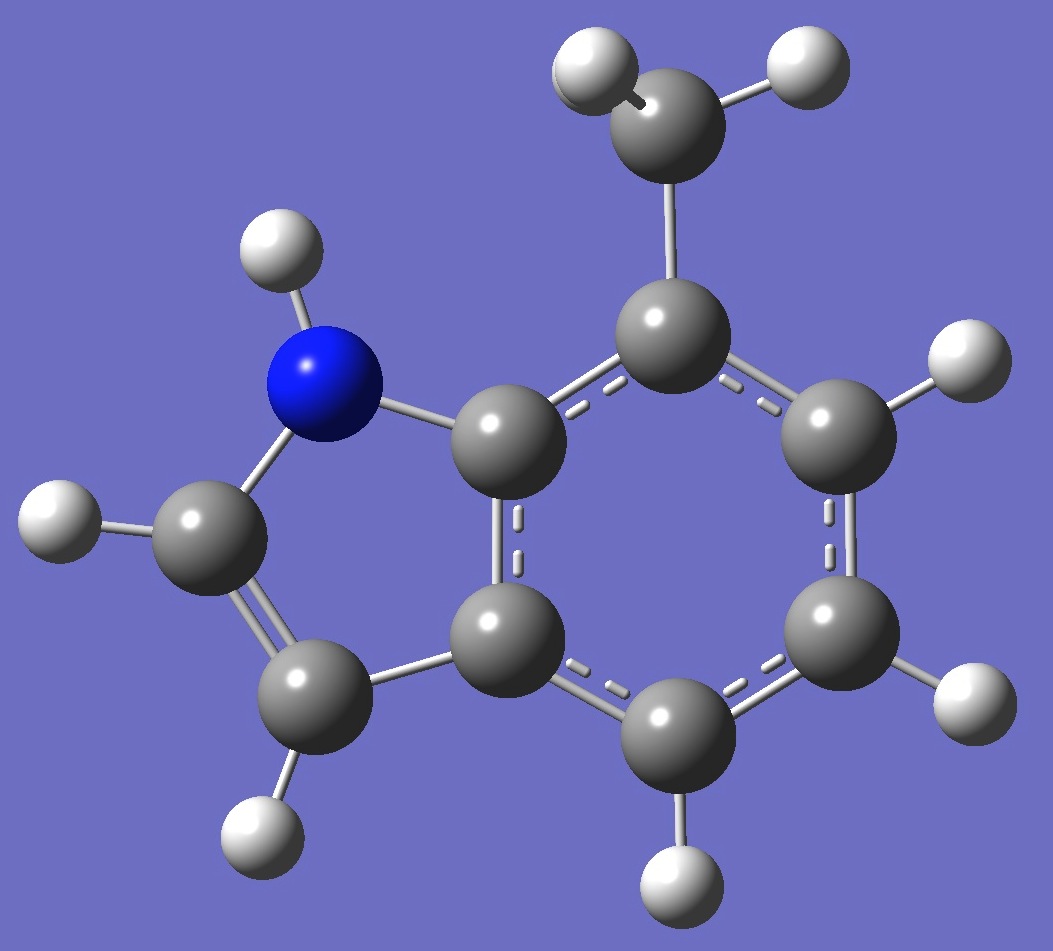
| Table 2. 7-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
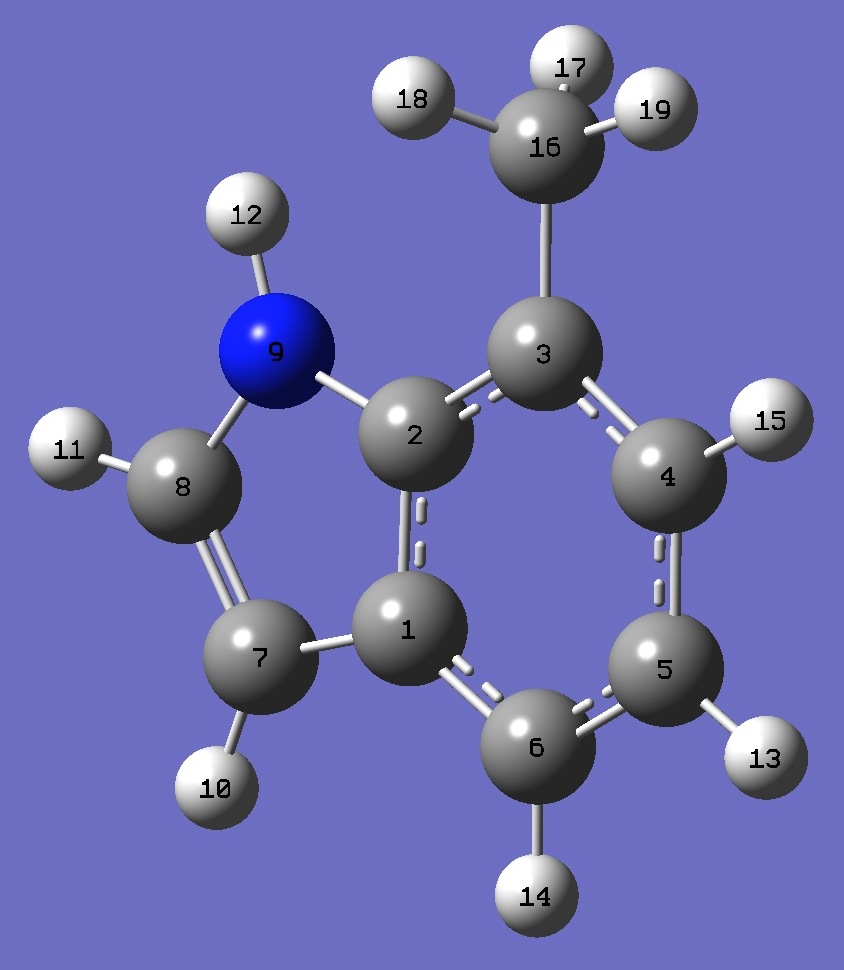
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,7,B9,1,A8,6,D7,0
H,8,B10,7,A9,1,D8,0
H,9,B11,2,A10,1,D9,0
H,5,B12,4,A11,3,D10,0
H,6,B13,5,A12,4,D11,0
H,4,B14,3,A13,2,D12,0
C,3,B15,2,A14,1,D13,0
H,16,B16,3,A15,2,D14,0
H,16,B17,3,A16,2,D15,0
H,16,B18,3,A17,2,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41670127
B2=1.39953528
B3=1.38667548
B4=1.40571512
B5=1.38100875
B6=1.43304838
B7=1.36473838
B8=1.37821327
B9=1.07653747
B10=1.07655071
B11=1.00274363
B12=1.08186744
B13=1.08211169
B14=1.08305963
B15=1.50330862
B16=1.0940714
B17=1.0940714
B18=1.08895359
A1=123.22748768
A2=115.87907917
A3=122.18041788
A4=121.23399686
A5=134.27439992
A6=107.17176942
A7=107.17644694
A8=127.02057639
A9=129.98241136
A10=125.61897187
A11=118.9849785
A12=120.75146842
A13=118.77984126
A14=121.39066959
A15=111.63133037
A16=111.63133037
A17=111.01908452
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=0.
D8=180.
D9=180.
D10=180.
D11=180.
D12=180.
D13=180.
D14=-59.97200271
D15=59.97200271
D16=180.
|
B1=1.41618084
B2=1.39882124
B3=1.38674475
B4=1.40525137
B5=1.3814704
B6=1.43150593
B7=1.36605018
B8=1.37524861
B9=1.07958664
B10=1.07975386
B11=1.0048377
B12=1.08495219
B13=1.08505073
B14=1.08619517
B15=1.49892725
B16=1.09638098
B17=1.09638098
B18=1.09141664
A1=123.37757826
A2=115.81088934
A3=122.17112727
A4=121.27277282
A5=134.38668151
A6=107.07199383
A7=107.2344235
A8=127.14203481
A9=130.13014216
A10=125.44529582
A11=118.99032034
A12=120.75822619
A13=118.65695391
A14=121.40690621
A15=111.69580839
A16=111.69580839
A17=110.93753463
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=0.
D8=180.
D9=180.
D10=180.
D11=180.
D12=180.
D13=180.
D14=-60.03188165
D15=60.03188165
D16=180.
|
|
|
|
|