|
| |
|
|
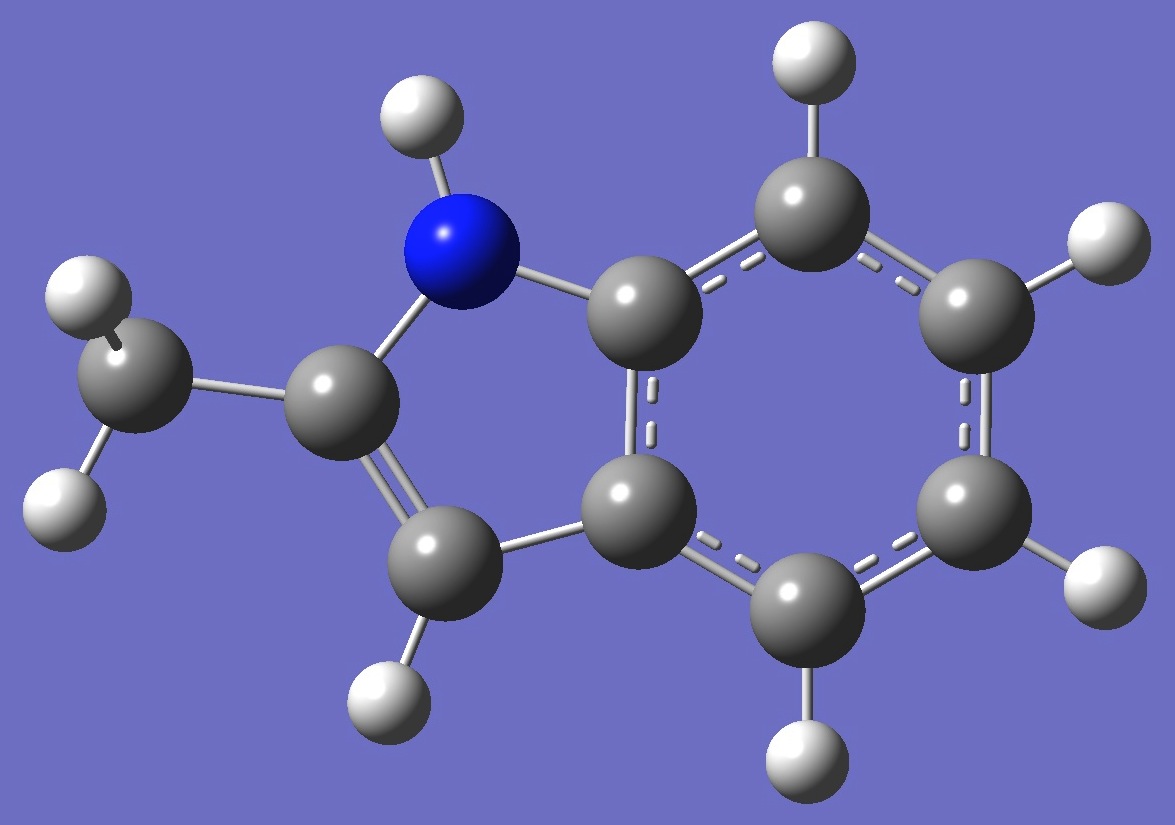
| Table 2. 2-Methylindole. Optimized molecular structure parameters, ropt (Å
and degrees). |
| |
|
|
B3LYP/cc-pVTZ and B3P86/6-31G(3d,3p)
|
| |
|
|
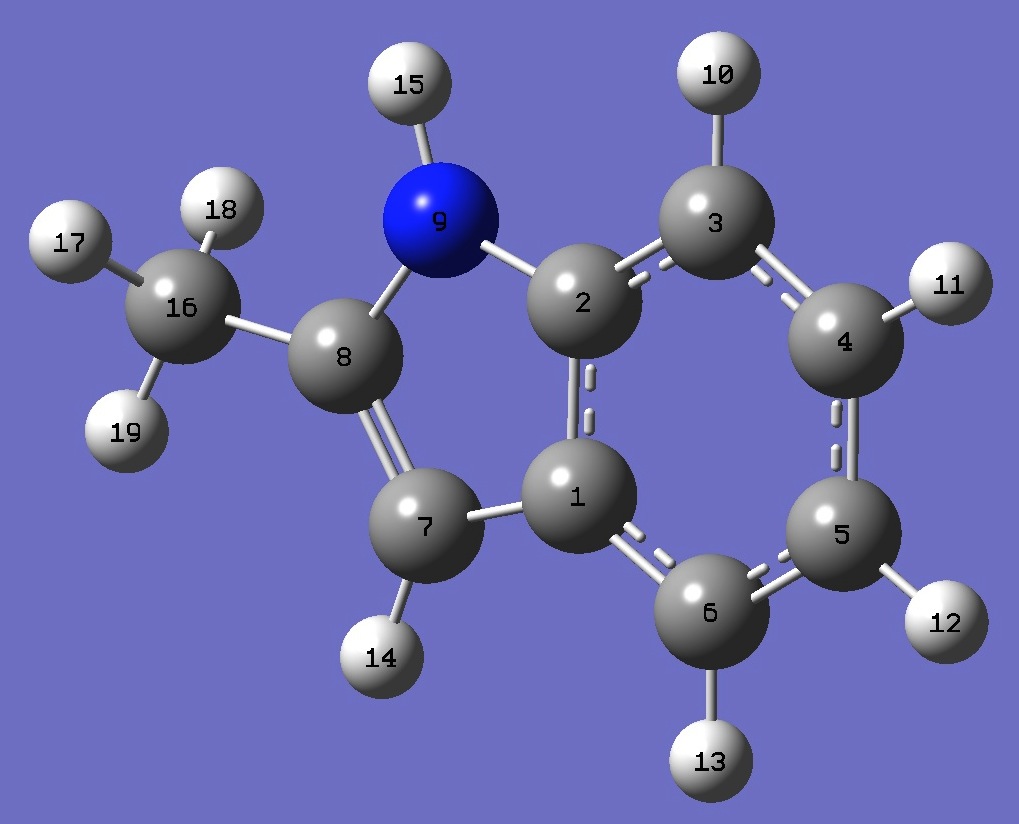
|
C
C,1,B1
C,2,B2,1,A1
C,3,B3,2,A2,1,D1,0
C,4,B4,3,A3,2,D2,0
C,5,B5,4,A4,3,D3,0
C,1,B6,6,A5,5,D4,0
C,7,B7,1,A6,6,D5,0
N,2,B8,1,A7,6,D6,0
H,3,B9,2,A8,1,D7,0
H,4,B10,3,A9,2,D8,0
H,5,B11,4,A10,3,D9,0
H,6,B12,5,A11,4,D10,0
H,7,B13,1,A12,6,D11,0
H,9,B14,2,A13,1,D12,0
C,8,B15,7,A14,1,D13,0
H,16,B16,8,A15,7,D14,0
H,16,B17,8,A16,7,D15,0
H,16,B18,8,A17,7,D16,0
|
|
|
B3LYP
|
B3P86 |
|
|
B1=1.41763846
B2=1.39201945
B3=1.38582293
B4=1.40372442
B5=1.38420653
B6=1.43353383
B7=1.367588
B8=1.37820966
B9=1.08250307
B10=1.08172812
B11=1.08183338
B12=1.08240385
B13=1.07707892
B14=1.00366679
B15=1.48966932
B16=1.09344023
B17=1.09344023
B18=1.0880166
A1=122.30846539
A2=117.56426432
A3=121.12756287
A4=121.11831628
A5=134.52387425
A6=107.80143532
A7=107.01477976
A8=121.44016278
A9=119.43175635
A10=119.1862736
A11=120.46449139
A12=126.9019671
A13=125.29929244
A14=130.38746536
A15=111.67171181
A16=111.67171181
A17=110.13782167
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=0.
D12=180.
D13=180.
D14=119.8376394
D15=-119.8376394
D16=0.
|
B1=1.41758756
B2=1.39206116
B3=1.3856695
B4=1.40357559
B5=1.38417346
B6=1.43139519
B7=1.36883298
B8=1.37502923
B9=1.08537312
B10=1.08481225
B11=1.08489446
B12=1.08532266
B13=1.08015147
B14=1.0054906
B15=1.48641009
B16=1.09553869
B17=1.09553869
B18=1.09029993
A1=122.43432839
A2=117.42077156
A3=121.19804969
A4=121.16765778
A5=134.5319179
A6=107.66210557
A7=107.05462413
A8=121.49847894
A9=119.37995462
A10=119.17692234
A11=120.50989008
A12=127.10469023
A13=125.27931911
A14=130.35975629
A15=111.76191149
A16=111.76191149
A17=109.98136492
D1=0.
D2=0.
D3=0.
D4=180.
D5=180.
D6=180.
D7=180.
D8=180.
D9=180.
D10=180.
D11=0.
D12=180.
D13=180.
D14=119.76054181
D15=-119.76054181
D16=0.
|
|
|
|
|